This web page was produced as an assignment for an undergraduate course at Davidson College.
BLM gene encodes a RecQ Helicase
What is BLM?
The BLM gene encodes a protein called RecQL3 helicase, which is a member of
the RecQ helicase family. Mutations in the BLM gene are responsible for Bloom’s
Syndrome, an autosomal recessive disease characterized by high levels of sister
chromatid exchange. The BLM gene was positionally cloned in 1995 and is located
on the long arm of Chromosome 15 (map locus 15q26.1) (Ellis, 1995).
RecQ Family
The RecQ family of helicases are enzymes that unwind DNA so that replication,
transcription, and DNA repair can occur. Helicases are vital to the life of
a cell and aid in the maintenance of genomic stability. (See Figure
1 for a cartoon of DNA helicase activity.) There are five known human
RecQ helicases which are RECQL, WRN, BLM, RECQL4, and RECQL5. Mutations in WRN
and RECQ4 cause Werner’s Syndrome and Rothmund-Thomson Syndrome , respectively.
Both of these diseases, along with Bloom’s Syndrome, increase an individual’s
predisposition to cancer (Karow, 2000). The RecQ helicases were named after
the E. coli RecQ helicase homologues and share similar recombination pathways.
All RecQ helicases share conserved domains (Luo, 2000).

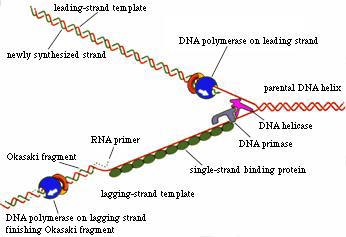
Figure 1. One function of DNA helicase is to unwind the double-stranded DNA at the replication fork so that DNA polymerase can replicate the DNA strands. (Permission pending for image found at http://sun0.mpimf-heidelberg.mpg.de/History/Hoffmann.html.)
Structure
The BLM protein is a ring helicase; it forms symmetric oligomeric rings similar
to other E. coli ring helicases (Karow, 2000). BLM mRNA is 4437 kB in length
and encodes a protein of 1417 amino acids with an estimated molecular weight
of 159 kDa that contains seven different domains. The domains include a poly-aspatartate
amino terminus domain (PD1), a poly-serine domain (PS), a poly-aspartate domain
(PD2), DEAH helicase domain (DEAH), RecQ helicase C-terminus domain (RecQCt),
helicase and RNase D C-terminus domain (HRDC), and nuclear localization signals
(NLS) (IMT Bioinformatics, http://bioinf.uta.fi/BLMbase/blmintro.html).
For the complete DNA and amino acid sequence of BLM, click here
and go to Bioimformatics. See Figure 2 for a representation
of the BLM domains.
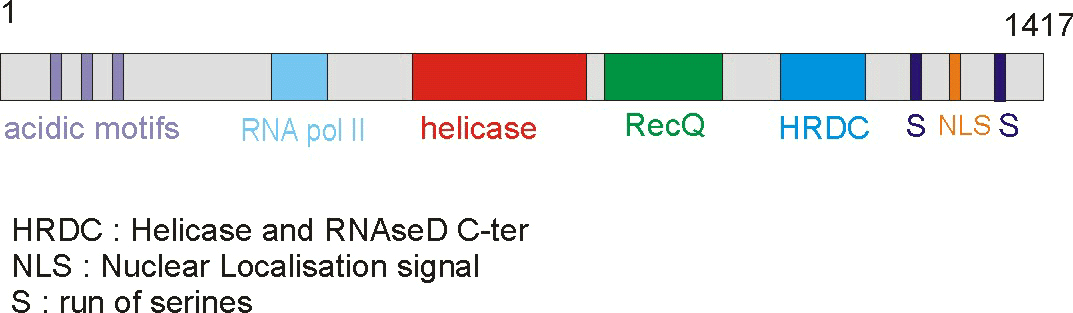
Figure 2. This cartoon depicts the BLM gene with its distinct domains. The helicase domain is conserved throughout different species. (Permission pending for image found at http://www-cryst.bioc.cam.ac.uk/~fabien/Structural_study_of_blm.html.)
Location
The gene product of BLM is primarily found in the nucleus of cells in nuclear
domain 10 (ND10), also known as promyelocytic leukemia (PLM). The level of BLM
in a cell fluctuates throughout the cell cycle. There are high amounts of BLM
in S phase and the levels remain high during G2 and M phase. Levels of BLM greatly
decrease during G1 phase. In S phase, the protein is localized in the nucleolus.
BLM is most likely involved in a DNA surveillance mechanism during S phase and
ND10 helps to regulate BLM levels during the cell cycle (Enomoto, 2001).
Function
Like all of the RecQ helicases, BLM unwinds double-stranded DNA in a 3’
to 5’ direction and requires ATP. BLM is also able to unwind G-quadruplex
DNA (G4 DNA), a highly stable structure of four guanines. G4 DNA must be unwound
in order for DNA metabolism to occur, and the RecQ helicases are the only enzymes
capable of such a function (Karow, 2000).
BLM is believed to interact with topoisomerase III (topo III). Topo III has an effect on the degree of DNA supercoiling during replication and transcription and aides in untangling daughter chromosomes at the end of S-phase so that the sister chromatids can segregate during mitosis. It is also thought that BLM suppresses homologous recombination, as Bloom Syndrome cells (containing a mutation in the Bloom gene) show highly increased recombination. So basically, the hypothesized roles for BLM, in addition to 3' to 5' helicase activity, include disruption of G4 DNA, disrupstion of sister chromatid recombination, and disruption of joint molecules (Karow, 2000).
Homologues
The human RecQ helicases all share a common helicase domain with other RecQ
helicases. The sequence has been highly conserved across species. Other organisms
that have been identified with homologous RecQ helicase sequences include bacteria
(E. coli), fungi (S. cerevisiae and S. pombe), plants
(Arabidopsis), and animals (Drosophila and C. elegans).
While BLM has a conserved helicase domain, it has an extended amino-terminus
and carboxyl terminus domain (Karow, 2000). See Figure 3 for
a comparison of the RecQ helicases in different species.
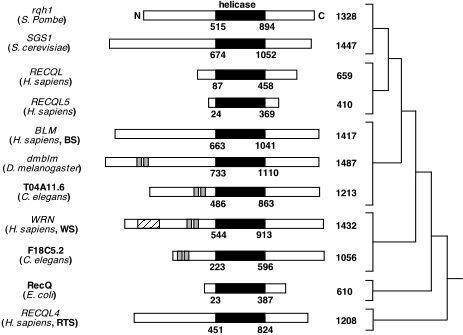
Figure 3. This image displays the family of RecQ helicases. The conserved helicase domain is depicted in black. (Permission pending for image found at http://www.hms.harvard.edu/pathol/sinclair/Pages/recqfamily.html.)
Bloom Syndrome
Bloom Syndrome is a rare autosomal recessive genetic disease due to mutations
on the BLM gene. Symptoms include small body size, sunlight sensitivity, immunodeficiency,
male infertility, hypo/hyper-pigmented skin, and a predisposition to malignant
tumors (Ellis, 1995). Individuals with Bloom Syndrome are more prone to cancer,
diabetes, and chronic lung disease. This disease is most common among the Ashkenazi
Jewish population, where one of every 100 Jews is a carrier (InTouchLive.com,
http://www.intouchlive.com/home/frames.htm?http://www.intouchlive.com/cancergenetics/bloom.htm&3).
Cells of Bloom Syndrome patients exhibit great genomic instability due to chromosome
breaks, homologous recombination, and sister chromatid exchange (SCE). Individuals
of the disease must be more aware of cancerous tumors and have the least amount
of exposure possible to the sun and X-rays. See Figure 4 for
an example of symtoms of a Bloom Syndrome patient.
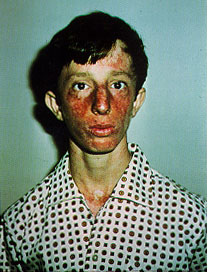
Figure 4. This is a picture of a patient with Bloom's Syndrome exhibiting the characteristic symptoms including sunlight sensitivity and discolored skin. (Permission pending for image found at http://web.mit.edu/biology/guarente/human/human.html.)
Allelic Variants
Several different mutations in the BLM gene have been identified as causing
Bloom Syndrome. In four unrelated Ashkenazi Jews, a 6 bp deletion/7 bp insertion
was discovered that resulted in an amino acid change and a premature stop codon.
Another mutation was identified in a Japanes patient such that one codon was
deleted resulting in a premature stop codon. A third mutation found in an Italian
patient involved a T to C transition that changed isoleucine to threonine (OMIM,
http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=604610).
Related Sites
http://www.mazornet.com/genetics/bloom_syndrome.asp
This site provides information on Bloom’s Syndrome along with other genetic
diseases found primarily in the Jewish population.
http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=604610
This site from OMIM contains information about RecQ helicases and Bloom’s
Disease. It also has links to relevant scientific papers.
http://opbs.okstate.edu/%7Emelcher/MG/MGW1/MG1223.html
Visit this site to learn more about SCE (sister chromatid exchange).
__________________________________________________________________________
Davidson College Biology Department
Davidson College Molecular Biology
Send questions and comments to Sarah Baxter at sabaxter@davidson.edu. Thanks!