This
page was produced as an assignment for an undergraduate course at
Davidson College.

Single-Strand Conformation Polymorphism
(SSCP)
DEFINITION
- SSCP is the electrophoretic separation of single-stranded nucleic acids based on subtle differences in sequence (often a single base pair) which results in a different secondary structure and a measurable difference in mobility through a gel.
BACKGROUND
- The mobility of double-stranded DNA in gel electrophoresis is dependent on strand size and length but is relatively independent of the particular nucleotide sequence. The mobility of single strands, however, is noticeably affected by very small changes in sequence, possibly one changed nucleotide out of several hundred. Small changes are noticeable because of the relatively unstable nature of single-stranded DNA; in the absence of a complementary strand, the single strand may experience intrastrand base pairing, resulting in loops and folds that give the single strand a unique 3D structure, regardless of its length. A single nucleotide change could dramatically affect the strand's mobility through a gel by altering the intrastrand base pairing and its resulting 3D conformation (Melcher, 2000).
- Single-strand conformation polymorphism analysis takes advantage of this quality of single-stranded DNA. First announced in 1989 as a new means of detecting DNA polymorphisms, or sequence variations, SSCP analysis offers an inexpensive, convenient, and sensitive method for determining genetic variation (Sunnucks et al., 2000).
- Like restriction fragment length polymorphisms (RFLPs), SSCPs are allelic variants of inherited, genetic traits that can be used as genetic markers. Unlike RFLP analysis, however, SSCP analysis can detect DNA polymorphisms and mutations at multiple places in DNA fragments (Orita et al., 1989). As a mutation scanning technique, though, SSCP is more often used to analyze the polymorphisms at single loci, especially when used for medical diagnoses (Sunnucks et al., 2000).
SSCP PROCEDURE
- The procedure used during the development of SSCP was as follows:
- digestion of genomic DNA with restriction endonucleases
- denaturation in an alkaline (basic) solution
- electrophoresis on a neutral polyacrylamide gel
- transfer to a nylon membrane
- hybridization with either DNA fragments or more clearly with RNA copies synthesized on each strand as probes (Orita et al., 1989).
- Since then, more convenient procedures have been developed, taking into account other molecular techniques, although sometimes it is simpler to amplify the double strand and then denature it into single strands instead of trying to find suitable primers for the below PCR method if the targeted sequence is unknown.
- Most experiments involving SSCP are designed to evaluate polymorphisms at single loci and compare the results from different individuals.
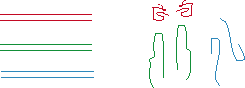 A
A
 B
B
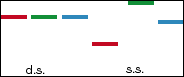 C
C
Figure 1: SSCP Procedure. The three equal-length double-stranded DNA
fragments are shown with the corresponding single-stranded structures, the red
fragment folding into the smallest molecule and the green the largest (Panel
A). The desired polymorphism is selected with PCR primers; primer A is in
excess to amplify only a single strand (Panel B). Both the
double-stranded and single-stranded fragments are run through gel
electrophoresis (Panel C). If not for the color labels, there would be no
distinction between the double-stranded fragments. The single-stranded
fragments, however, show considerable variation in mobility; the small red
molecule migrates more quickly through the gel than either the blue or the
large green molecule. Using this SSCP result, it becomes clear that the
different lanes (red, blue, or green) contain strands with different sequences;
the more far apart the bands, the less similar the nucleotide sequences.
Source with Permission from Dr. Ulrich
Melcher
- Procedure as illustrated in Figure 1:
1. A specific pair of PCR primers (forward and
reverse) is used to amplify the desired DNA fragments from individuals.
2.
Single-stranded DNA is produced by asymmetric PCR: the primer on one side
of the fragment is greatly in excess over the other primer. After the
low-concentration primer supply is
exhausted, continued PCR produces only the target single strand.
3.
The mobilities of the single stranded fragments are compared by
electrophoresis on a neutral polyacrylamide gel.
4. Bands
are detected by radioactive labeling or (more often) silver staining, and the
pattern is interpreted (Melcher, 2000).
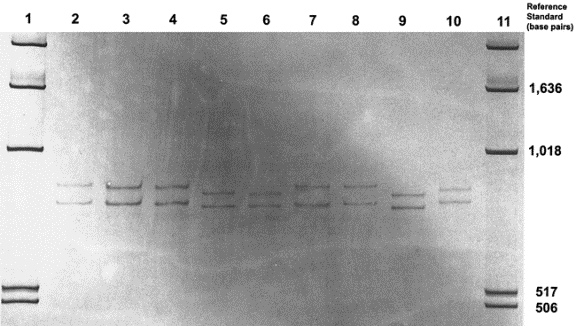
Figure 2: Sample SSCP Gel Result and Interpretation.
DNA was isolated and amplified from sand flies (Lutzomyia longipalpis).
SCCP analysis of the DNA shows multiple haplotypes, or sets of alleles
usually inherited as a unit. Lanes 3 and 4 were identical haplotypes from
two individuals. The difference in band migration in adjacent lanes is
associated with the number of nucleotide differences (in parentheses): lanes
2-3 (2), lanes 3-4 (0), lanes 4-5 (3), lanes 5-6 (1), lanes
6-7 (3), lanes 7-8 (1), lanes 8-9 (1), and lanes 9-10 (4).
Source: Hodgkinson,
et al,. 2002
SSCP LIMITATIONS AND CONSIDERATIONS
- Single-stranded DNA mobilities are dependent on temperature. For best results, gel electrophoresis must be run in a constant temperature.
- Sensitivity of SSCP is affected by pH. Double-stranded DNA fragments are usually denatured by exposure to basic conditions: a high pH. Kukita et al. found that adding glycerol to the polyacrylamide gel lowers the pH of the electrophoresis buffer--more specifically, the Tris-borate buffer--and the result is increased SSCP sensitivity and clearer data (1997).
- Fragment length also affects SSCP analysis. For optimal results, DNA fragment size should fall within the range of 150 to 300 bp, although SSCP analysis of RNA allows for a larger fragment size (Wagner, 2002). Tthe presence of glycerol in the gel may also allow a larger DNA fragment size at acceptable sensitivity (Kukita et al., 1997).
- Under optimal conditions, approximately 80 to 90% of the potential base exchanges are detectable by SSCP (Wagner, 2002).
- If the specific nucleotide responsible for the mobility difference is known, a similar technique called Single Nucleotide Polymorphism (SNP) may be applied.
REFERENCES
- Hodgkinson, Virginia, et al. Rapid Identification of Mitochondrial Cytochrome B Haplotypes by SSCP in Lutzomyia longipalpis Populations. <http://www.bioone.org/bioone/?request=get-document&issn=0022-2585&volume=039&issue=04&page=0689>. Accessed 2003 February 17.
- Kukita, Y., et al. SSCP Analysis of Long DNA Fragments in Low pH Gel. Human Mutation; 1997, (10): 400-7.
- Melcher, Ulrich. SSCPs. <http://opbs.okstate.edu/~melcher/MG/MGW1/MG11129.html>. Accessed 2003 February 17.
- Orita, M., et al. Detection of Polymorphisms of Human DNA by Gel Electrophoresis as SSCPs. Proceedings of the National Academy of Sciences of the United States of America; 1989, (86): 2766-70.
- Sunnucks, P., et al. SSCP Is Not So Difficult: The Application and Utility of Single-Stranded Conformation Polymorphism in Evolutionary Biology and Molecular Ecology. Molecular Ecology; 2000, (9): 1699-710.
- Wagner, John. Screening Methods for Detection of Unknown Point Mutations. <http://www-users.med.cornell.edu/~jawagne/screening_for_mutations.html#Single-Strand.Conformational.Polymorphism>. Accessed 2003 February 17.
Molecular
Biology Homepage
Sarah's
Molecular Homepage
Email Sarah