This webpage was produced as an assigment for an undergraduate course in Molecular Biology at Davidson College.
Orthologs of Triosephosphate Isomerase
The crystal structure of Triosephosphate Isomerase (TIM) illustrated in the previous web assignment was obtained from the worm Caenorhabditis elegans. This webpage illustrates the orthologs of TIM in other common species such as humans (Homosapiens sapiens), mouse (Mus musculus), fruitfly (Drosophila melanogaster), Arabidopsis (Arabidopsis thaliana), yeast (Saccharomyces cerivisiae), and Escherechia coli.
A search on wormbase yielded a picture of the TIM gene on Chromosome II in C. elegans (Figure 1).
![]()
Figure 1. Picture of the Triosephosphate Isomerase (tpi-1) gene in C. elegans. Introns are regions of the chromosome represented by a thin, black line and exons are represented by green boxes. (Wormbase).
Moreover, a nucleotide sequence was obtained from NCBI showing 2387 bases in the gene (Figure 2). This sequence includes both introns and exons.

Figure 2. Nucleotide base sequence of tpi-1. (NCBI)
A Blastn search of the above sequence against genomes of all organisms gave only a long list of TIM clones in C. elegans rather than showing sequence homology with other organisms. Individual Blastn searches of the tpi-1 gene with certain organisms gave E-values for each (Figure 3). While Mus musculus appeared to have the highest E-value of 0.089 i.e. highest degree of sequence homology with C. elegans, Drosophila had the lowest E-value of 0.0008 and therefore had the least sequence homology with the tpi-1 gene in C. elegans. Although searches on NCBI indicated the presence of a tpi-1 gene in humans, rats, yeast and E. coli, a Blastn stated that there was no significant sequence similarity between the tpi-1 gene in these organisms and C. elegans.

Figure 3. Blastn results from individual Blastn searches of C. elegans against genomes of other organisms. Rows from top to bottom represent the following organisms: Arabidopsis, Drosophila, Chicken, and mouse. The column on the far right represents E-values. (NCBI Blastn).
The amino acid sequence for triosephosphate isomerase shows that it has 247 residues (Figure 4). Moreover, its molecular weight is approximately 26.6 kDa and it has a isoelectric point (pI) of 6.68 (Wormbase).

Figure 4. Amino acid sequence for Triosephosphate Isomerase. (Wormbase).
The above amino acid sequence was used to perform a Blastp search. This search yielded several hits in common species but with low E-values (Figure 5). Arabidopsis TIM appears to have the relative highest homology of amino acid sequence to C. elegans as indicated by the E-value of 2 e-65, and this corroborates the previous observation that the TIM in Arabidopsis has high homology of base sequence determined by Blastn with C. elegans. Chicken TIM, on the other hand, appears to have least homology of amino acid sequence to C. elegans TIM. However, although TIM appears to perform an important function in the glycolytic pathway and C. elegans TIM does have orthologs in other organisms, these orthologs do not share a high degree of homology.

Figure 5. Blastp results from using amino acid sequence of TIM from C. elegans versus other organisms. Rows from top to bottom represent the following organisms: Chicken, Anopheles gambiae, Drosophila, Human, Mouse, Rat, Arabidopsis, Arabidopsis, and Yeast. The column on the far right represents E-values. (NCBI Blastp).
The ENSEMBL website yielded a table of C. elegans TIM orthologs found in other organisms. As per these alignments, these TIM proteins in other organisms have a good degree of homology with C. elegans TIM (Figure 6).
(a) Alignment of TIM sequence in Homosapiens sapiens with C. elegans TIM.

(b) Alignment of TIM sequence in Gallus gallus with C. elegans TIM.

(c) Alignment of TIM sequence in Mus musculus with C. elegans TIM.

(d) Alignment of TIM sequence in Saccharomyces cerivisiae with C. elegans TIM.

(e) Alignment of TIM sequence in Drosophila melanogaster with C. elegans TIM.

Figure 6. Alignments of TIM amino acid sequences in (a) Humans (Homosapiens sapiens); (b) Chicken (Gallus gallus); (c) Mouse (Mus musculus); (d) Yeast (Saccharomyces cerivisiae); and (e) Fruitfly (Drosophila melanogaster) with TIM sequence for C. elegans. (ENSEMBL).
A conserved domain search using NCBI rpsblast showed that TIM has domains similar to a bacterial protein phosphoglycerate kinase (Figure 6). It also has domains that are conserved in 636 other TIM sequences (Figure 7), thereby indicating a large number of orthologous proteins.
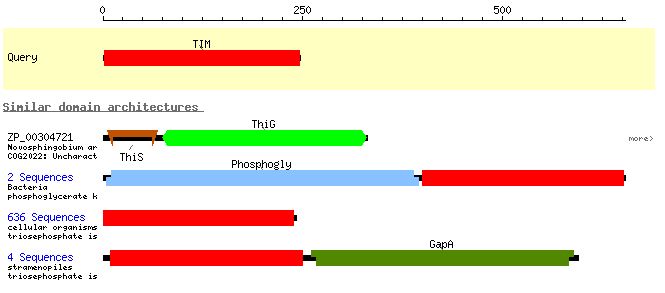
Figure 7. Conserved domains between the C. elegans TIM protein and orthologous proteins in other organisms. The red domain indicated the TIM domain that is seen in other proteins as well. (NCBI Conserved Domain Database rpsblast)
Thus, while the C. elegans TIM appears to have conserved domains with other proteins, the Blastn and Blastp results point towards a low degree of sequence homology.
References:
Wormbase. <http://www.wormbase.org/>. Accessed 7th March, 2005.
National Center for Biotechnology Information. <http://www.ncbi.nlm.nih.gov/>. Accessed 7th March, 2005.
European Molecular Biology Laboratory-European Bioinformatics Institute, and Sanger Institute. ENSEMBL. <http://www.ensembl.org/>. Accessed 7th March, 2005.
Nupur's Molecular Biology Homepage