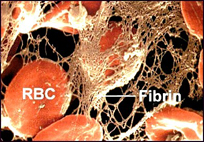

A thrombus is simply another word for clot; in this case, a blood clot (Figure 1). Thrombi are formed in response to damage to the wall of a blood vessel under normal conditions. This prevents the loss of blood through the damaged portion of the vessel. The pathogenic form is called thrombosis and occurs when a clot forms in an area that is undamaged or when a naturally-occurring clot is transported to a blood vessel that is blocked by its presence.
Thrombin is a multi-functional serine protease that is involved in a number of activitives within the body (While we will touch upon many of its functions, we will focus primarily upon its ability to cleave fibrinogen macromolecules into fibrin monomers). The crystal structure of human a-thrombin was determined in 1991 by Bode et al. Thrombin shares many similarities with other serine proteases such as trypsin, which is found in the stomach of many organisms and aids in the non-specific breakdown of proteins that are consumed by the organism. Where thrombin differs from other members of its family is in its specificity. Its production is triggered near the end of the coagulation cascade in response to injury that requires the formation of a blood clot. Within this pathway, its main function is to cleave fibrinogen into fibrin molecules, which form the bulk of the clot or thrombus. Like trypsin, thrombin prefers cleaving behind either arginine or lysine amino acids, but preferentially cleaves Arg-Xaa bonds( Xaa simply stands for any other amino acid attached to Arg). Even within fibrinogen molecules, it only cleaves two Arg-Xaa bonds within the macromolecule(Bode et al, 1992).
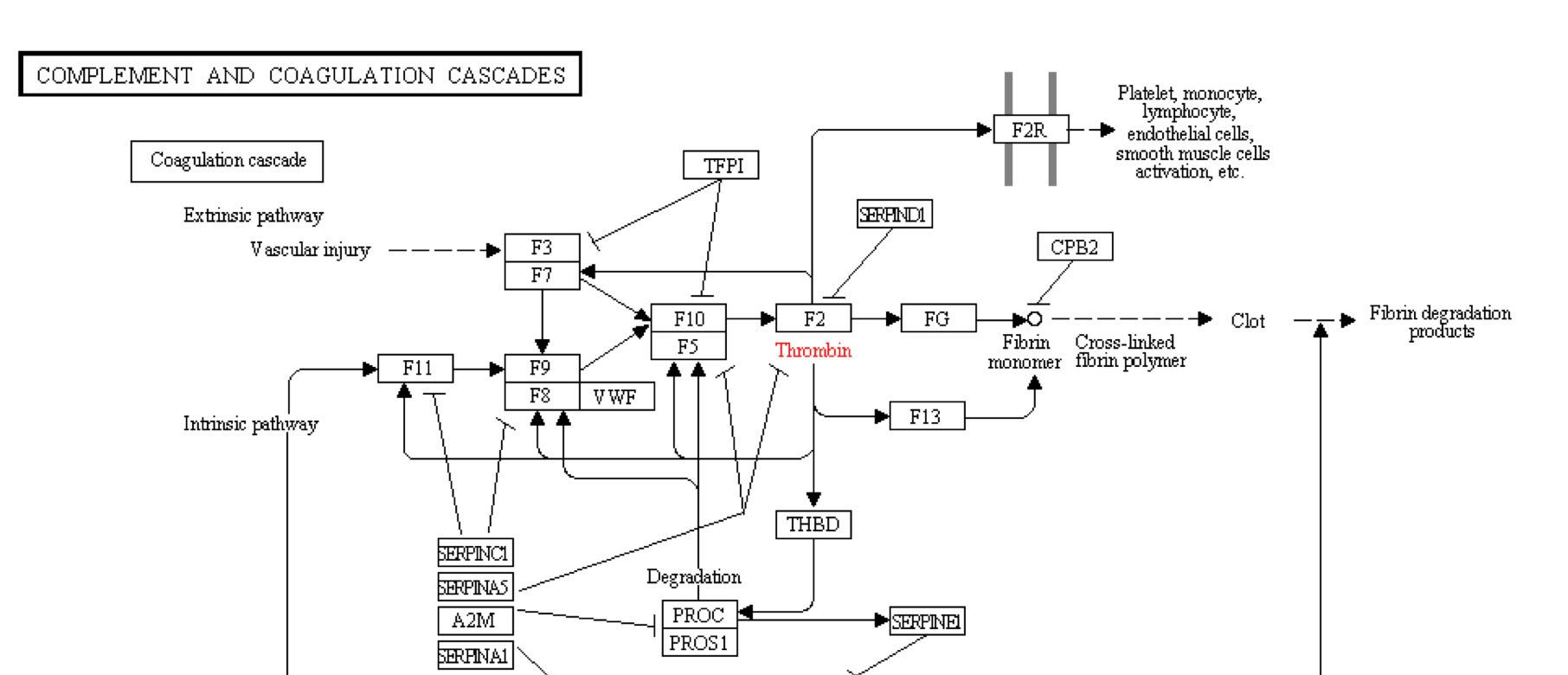
While thrombin’s main function is producing fibrin monomers, it is integrated with the rest of the coagulation cascade as shown by the KEGG coagulation pathway in Figure 2.Thrombin also activates other proteins in the coagulation cascade such as coagulation factors II, IV, XIII, XI and XIII (Figure 2). Its activation of other coagulation factors act as a positive feedback loop affecting many of the proteins involved in the clotting process, as well as thrombin production. When bound to thrombomodulin it can also enhance Protein C capabilities but loses its proteolytic capacity while bound. Thrombin’s activation of Protein C also leads to activation of smooth muscle cells, endothelial cells, lymphocytes, monocytes and platelets (Bode et al, 1992).
Thrombin begins life as a more complex and longer protein known as thrombinogen or--more commonly--prothrombin The composition of prothrombin versus thrombin is shown in Figure 3. The B chain in the final thrombin protein is constructed from the 259 aa at the carboxyl end of the prothrombin protein. Prothrombin is a vitamin-K dependent zymogen. Vitamin K is necessary for it to assume its final three-dimensional shape due to a large number of glutamate residue. These Gla amino acids allow prothrombin to bind calcium and adhere to phospholipid bilayers. Once bound, it is possible for factors Xa and Va to bind and form the prothrombinase complex in preparation for proteolysis (Source).
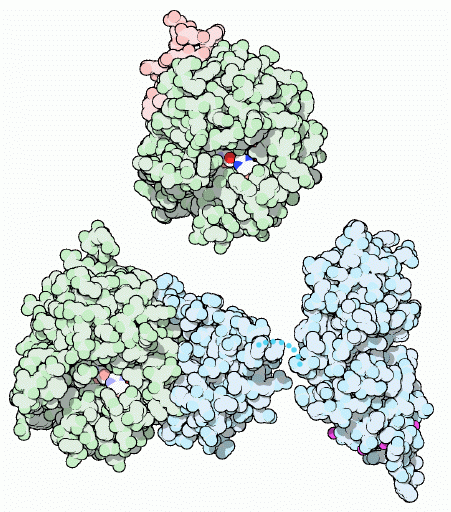
Shown in Figure 1 is the cleavage of prothrombin's amino end to leave behind the 259 amino acid carboxyl end (B Chain), which remains in the final thrombin molecule shown in the top portion of the figure. The pink section is the much shorter A Chain. The A Chain is located opposite from thrombin's active site and is thought to be involved with the cleavage of prothrombin's carboxyl end. Its distance from the active site makes in unlikely that any molecule associated with the active site would come into contact with the A chain at the opposite side of the much larger B Chain. It has also been shown that it is unecessry for thrombin's proteolytic ability(Bode et al, 1992). Unlike many proteins, the A and B chains do not form separate constructs within the final, quaternary structure, but rather form one, uneven and indented spherical protein (Bode et al, 1989).
Thrombin is peculiar, in that it contains a high number of polar and charged molecules that are distributed unequally over its length. More than 70% of all amino acids on the surface of thrombin are hydrophilic. 79 out of 89 charged amino acids are exposed on the surface of the molecule, while 10 are completely hidden from the external environment within the folds of the protein. Most of these charged residues are arranged in complementary pockets such that negatively and positively charged amino acids are bonded to one another. These interactions are particularly important in maintaining the bond between the A and B chains as well as the structuralintgrity of the protein as a whole. There are, however, three sections of the protein that are comprised of concentrated positively or negatively charged amino acids, which actually create instability within the molecule. Two opposing poles of the molecule show a net positive charge while a ring-like structure in the middle of the enzyme is comprised mainly of negatively charged amino acids. This final structure surrounds the active site of the thrombin molecule (Bode et al, 1992).
Thrombin works by cleaving two sites within the fibrinogen molecule after two arginine residue sites. When trypsin is exposed to fibrinogen, it cuts the macromolecule into approximately 150 fragments. Thrombin, however, will only cleave fibrinogen at two specific sites. It is thought that much of this specificity is due to the unequal spread of positively and negatively charged amino acids. A section of the a-fibrinogen molecule interacts with the negatively charged ring around the active site as well as the active site itself. Secondarily (but equally as importantly), one of the positively-charged exosites also interacts with the fibrinogen molecule. This is likely important in both orienting the fibrinogen molecule correctly within the active site and maintaining a strong bond between the enzyme and its substrate. Competitive inhibition of just the exosite incapacitates thrombin's ability to cleave fibrinogen (Fenton et al, 1991).

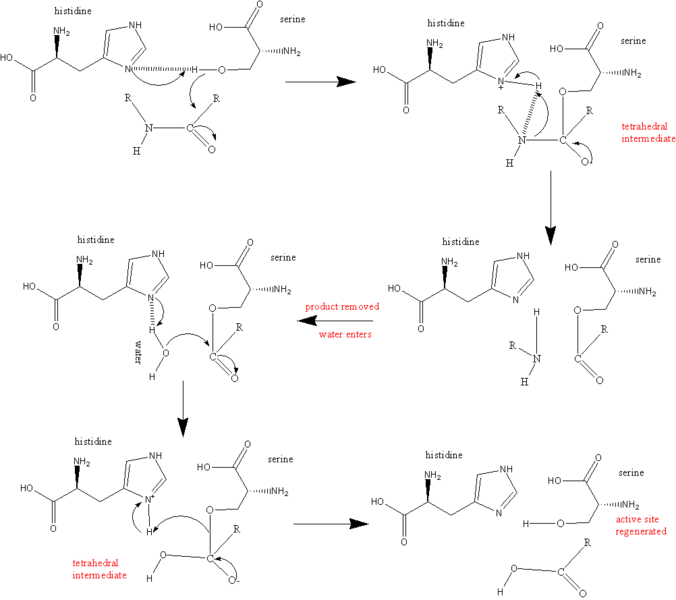
Thrombin's active site contains three amino acids also found in trypsin: Serine195, Aspartic acid102 and Histidine57. These three amino acids are responsible for recognizing and cleaving the proper sequences (Figure 4) (Martin et a, 1992). Upon recognition of the correct substrate sequence, the OH- groupfrom Ser195 cleaves the post-arginine bond (Source). It has also been demonstrated that thrombin shows equal affinity for fibrin as for fibrinogen, suggesting that the enzyme might bind to fibrin after it has been cleaved from fibrinogen (Bode et al, 1992). This is in keeping with papers that suggest that much of thrombin activity might occur late in the coagulation process from enzymes that have bound to already-cleaved fibrin molecules. It has been demonstrated that thrombin may be involved in repair of damaged blood vessels as well as clot formation (Schrorl, 2010).
When a substrate binds to thrombin, it must do so in a very precise way. The carbonyl end of the amino acid that is about to be cleaved from the substrate must be near the OH- group on the Ser195 thrombin amino acid. A nitrogen within His57 then acceptsa hydrogen atom from Ser195 while the Serine amino acid grabs hold of the carbon in the carbonyl end of the substrate's amino acid. The histidine also accepts a pair of electrons from the double bond that was originally formed between the carbon and oxygen in the cabonyl end of the substrate's amino acid. The peptide bond holding together the amino and carbonyl ends of the amino acid are now broken. The energy released from this breakage severs the hydrogen atom from His57 and attaches it to the amino end of the now-severed amino acid, which falls away from the active site. The electrons from the carbon-oxygen bond return to recreate the double-bond. Water (H2O) enters the active site and the oxygen binds to the carbon atom of the carbonyl of the cleaved amino acid, leaving the hydrogen atom attached to the nitrogen in H57. Finally, this hydrogen is reattached to the oxygen on Ser195 to reform its OH- group in preparation for the next substrate (Kraut, 1977). The whole process ofthe catalytic mechanism can be seen in Figure 5.
* All images taken from Wikipedia Commons unless otherwise noted.
Bode, W., Turk, D., Karshikov A. The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human a-thrombin: Structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. 1992. Protein Science. 426-471.
Bode, W., Mayr, I., Baumann, U., Stone, S. R., Hofsteenge, J. 1989. The Refined A crystal structure of human a-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. The EMBO Journal. 8(11): 3467-3475.
J.W. Fenton IIa, G.B. Villanuevab, F.A. Ofosuc, J.M. Maraganored. Thrombin Inhibition by Hirudin: How Hirudin Inhibits Thrombin. 1991. Haemostasis 1991;21 (Suppl. 1):27-31.
Karsten Schrör1; Ellen Bretschneider; Kerstin Fischer; Jens W. Fischer; Robert Pape; Bernhard H. Rauch; Anke C. Rosenkranz1;
Artur-Aron Weber. Thrombin receptors in vascular smooth muscle cells – function
and regulation by vasodilatory prostaglandins. 2010.
Kraut, J. 1977. Serine Proteases: Structure and Mechanism of Catalysis. 1977. Ann. Rev. Biochem. 46:331-58.
Martin, P. D., Robertson, W. Turk, D.,Huber, R., Bode, W. Edwards, B.F.P. 1992. The Structure of Residues 7-16 of the Aa-Chain of Human Fibrinogen Bound to Bovine Thrombin at 2.3-A Resolution. The Journal of Biological Chemistry. 267(11):7911-7920.
Please direct questions or comments to Sarah Pyfrom (sapyfrom@davidson.edu) or Dr. Malcolm Campbell (macampbell@davidson.edu).