*This website was produced as an assignment for an undergraduate course at Davidson College
α-Amylase Orthologs
What are orthologs?
Orthologs are defined as genes that have diverged after a speciation event (Fulton et al., 2006). Orthologs are typically more similar to each other than to other genes in the genome, which is why sequence similarity is often used to infer gene orthology between two or more species. It has also been found that orthologs tend to have similar function. As a result, they are very useful in comparative analyses. Overall, ortholog prediction is an important part of comparative genomics as it allows researchers to be able to further predict conservative regulatory genes upstream of orthologous genes.
Evolution of α-amylase:
Comparing sequences and structures of different α-amylases
As discussed in My Favorite Protein, α-amylase, specifically human α-amylase, is an enzyme that catalyzes the hydrolysis of α-1,4 glucan linkages in starch. Human α-amylase comes in two forms, salivary and pancreatic. Both forms share very similar primary sequences as well as function and are very closely linked. But α-amylase is found not only in humans but other organisms as well. Because starch ranks among the most abundant carbohydrate polymers on Earth, it is the most inportant source of energy for not only animals like humans, but higher plants and microorganisms as well (Janecek, 1997). Therefore, α-amylase is widely distributed in plants, mammalian tissues and microorganisms (Buisson et al., 1987) all having similar functions.
All α-amylases catalyze the hydrolysis of α-1,4-D-glucosidic bonds, yet their amino acid sequences have high variability (Janecek, 1994). Stefan Janecek found that although the amino acid sequences vary among different organisms, there are four small but well established regions that show a high degree of conservation. Janecek aligned whole amino acid sequences of three groups of α-amylases using CLUSTAL V. The three groups were the following: (a) fungi and yeasts, (b) plants, and (c) streptomycetes, insects and mammals. Some of his results are shown below:
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()

Figure 1. Sequence similarities in α-amylases. The abbreviations are as follows: Bacli = Bacillus licheniformis, Escco = Escherichia coli, Aspni = Aspergillus oryzae, Sacfi = Saccharomycopsis fibuligera, Barle = Hordeum vulgare (barley isozyme A), Maize = Zea mays (maize), Drome = Drosophila melanogaster, Muspa = Mus musculus (mouse pancreas), Pigpa = Sus scrofa (pig pancreas), Humpa = Homo sapiens (human pancrease), and Humsa = Homo sapiens (human saliva). The numbers represent the number of amino acid residues between two regions and the sizes of the peptide chain preceding the first and ending the last region. Invariable amino acid residues are indicated (*). Image courtesy of (Janecek, 1994). Permission pending.
The similarity, although only in short segments, is obvious throughout the set of α-amylases above, especially between members of the same group that Janecek clustered together (fungus and yeast, plants, and insects and mammals). The Bacillus licheniformis shares a very similar α-amylase sequence to E.coli, both common bacteria. The Aspergillus niger and Saccharomycopsis fibuligera share very similar α-amylase sequences between each other, both being members of the fungus and yeast group. Barley and maize, both plants, share similar α-amylase sequences as well. Finally, the fruit fly and mammal α-amylases show very similar amino acid sequences, with an identical first region. The regions represent different loops on the enzyme, dealing with the joining of a β- strand of the molecule with the adjacent α-helix. From previous studies, it is found that the different regions highlighted by Janecek are involved with the calcium ligand binding site and the catalytic area (Buisson et al., 1987).
To compare the entire amino acid sequence of α-amylase from different species to human α-amylase investigated under 'My Favorite Protein', a pairwise amino acid alignment was conducted using BLAST. The results for some species can be seen by clicking on the links below.
Human salivary a-amylase (56549660) vs. Extracellular Aspergillus niger a-amylase (CAK44871.1)
Human salivary a-amylase (56549660) vs. Maize a-amylase (ACF87219.1)
Human salivary a-amylase (56549660) vs. Drosophila melanogaster a-amylase (24654362)
Human salivary a-amylase (56549660) vs. Mouse pancreatic a-amylase (111607467)
**Human pancreatic a-amylase (4502085) vs. Proposed C.elegans a-amylase sequence (17558728)
With this knowledge of the sequence similarities in α-amylases, Janecek (1994) was able to create an evolutionary tree for all the different α-amylases. The unrooted tree is shown below.

Figure 2. A proposed evolutionary tree of α-amylases. The remaining abbreviations are as follows: Micsp = Micrococcus sp., Bacme = Bacillus megaterium, Salty = Salmonella typhimurium, Bacst = Bacillus stearothermophilus, Bacam = Bacillus amyloliquefaciens, Bacsu = Bacillus subtilis, Butfi = Butyrivibrio fibrosolvens, Xanca = Xanthomonas campestris, and Aerhy = Aeromonas hydrophila. The branch lengths are proportional to the divergency of the amino acid sequences of α-amylases, the sum of the lengths of the branches linking any α-amylases being a measure of the evolutionary distance between them. Image courtesy of (Janecek, 1994). Permission pending.
As mentioned before, human α-amylase has a characteristic (α/β)-barrel structure, specifically in Domain A leading to Domain B. This is seen as a common trait in all α-amylases. It can be suggested that when crystal structures of α-amylases from different species are compared, the similar amino acid sequences of the enzyme can be related to the (α/β)-barrel structure of the molecules, important in the active site of α-amylase.
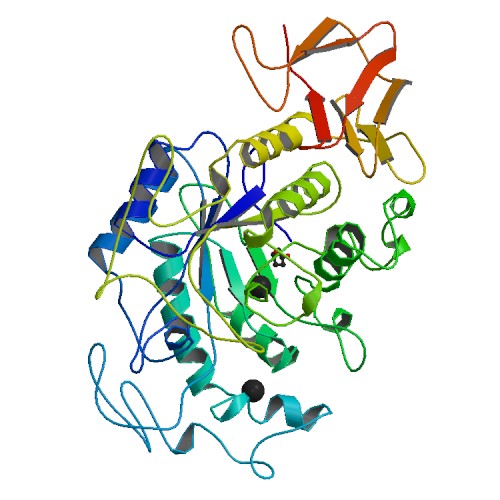
Figure 3. Crystal structure of human pancreatic α-amylase. Photo courtesy of PDB.
Figure 4. Crystal structure of pig pancreatic α-amylase bound to a substrate. Photo courtesy of PDB.
Figure 5. Crystal structure of Aspergillus niger α-amylase. Photo courtesy of PDB.
Figure 6. Crystal structure of barley α-amylase. Photo courtesy of PDB.
Figure 7. Crystal structure of Bacillus licheniformis α-amylase. Photo courtesy of PDB.
While none of the structures are exactly identical, they are all characterized by the (α/β)-barrel structure. This makes sense when looking at the aligned sequences in Figure 1, for none of the five structures shown have the same amino acid sequence even in the well established conservative regions. What is clear is that all the molecules have calcium ligands (indicated by the black balls), and appear to have similar structures around the calcium ions.
What do the comparisons mean?
All the similarities of both functional and sequence-structural nature reflect the very probable divergent evolution of α-amylase from a common ancestor (Janecek 1997). The ability to compare α-amylase amino acid sequences between species allows us to see the conserved sequences and deduce that those sequences are involved in the protein's function since all α-amylases perform a similar function, breaking down starch. The appearance of the calcium ion in the different α-amylase structures suggest that the calcium ion is very important in determining optimal enzymatic activity for all α-amylases. While the impact of the conserved regions on the function of various α-amylases is still difficult to define, the importance of the α-amylases is comparable with the importance of conserved regions (Janecek, 1994).
References:
Buisson G, Duee E, Haser R, Payan F. Three dimensional structure of porcine pancreatic α-amylase at 2.9 A resolution. Role of calcium in structure and activity. The EMBO Journal. 1987; 6: 3909-3916. PubMed.
Fulton DL, Li YY, Laird MR, Horsman BGS, Roche FM, Brinkman FSL. Improving the specificity of high-throughput ortholog prediction. BMC Bioinformatics. 2006; 7:270. <http://www.biomedcentral.com/1471-2105/7/270>
Janecek S. α-Amylase family: Molecular biology and evolution. Prog. Biophys. Molec. Biol. 1997; 67: 67-97. PubMed.
Janecek S. Sequence similarities and evolutionary relationships of microbial, plant and animal α-amylases. European Journal of Biochemistry. 1994; 224: 519-524. PubMed.
Please direct questions or comments to Abbey Webb