 How
to calculate the MW of a molecule that has been separated in a gel
How
to calculate the MW of a molecule that has been separated in a gelThe log of a molecule's molecular weight
is proportional
to the distance that molecule has migrated. Therefore, the first step is
to generate a standard curve using molecules of know size (the molecular
weight markers).
When using semilog paper (see below), the molecular weights (in
bp for DNA, and kiloDaltons (kDa) for proteins) is plotted on the Y-axis
and the distance the molecule migrated is plotted on the X-axis. When
generating
a standard "curve", you will obtain a straight line (use a best-fit
line).
Once your standard curve is ready, measure the distance traveled by your
molecule of interest. Find that distance on the X-axis, and go up until
you intersect with your standard curve. Move over to the Y-axis and that
will indicate the molecular weight of the molecule you are studying.
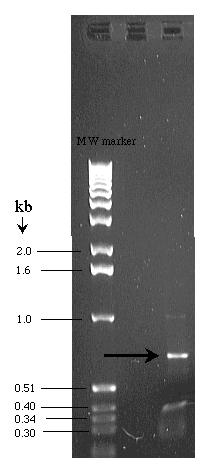
Use the graph paper below and the DNA gel shown to the right to
determine the molecular weight of the unknown band indicated with an
arrow.
Distance Migrated (mm)
Lab Schedule In Context of Research Project