Science . Vol. 265 . 1 July 1994
Hua Gu,* Jamey D. Marth, Paul C. Orban, Horst Mossmann, Klaus Rajewsky
H. Gu and K. Rajewsky Institute for Genetics, University
of Cologne, 50931 Cologne, Germany.
J. D. Marth, Biomedical Research Center and Department of Medical Genetics,
University of British Columbia, Vancouver, Canada V6T 1Z3.
P. C. Orban, Biomedical Research Center, University of British Columbia,
Vancouver, Canada V6T 1Z3.
H. Mossmann, Max-Planck Institute for Immunobiology, 79011 Freiburg, Germany.
*To whom corresnondence should be addressed.
7 April 1994; accepted 13 May 1994
Deletion of the promoter and the first exon of the DNA polymerase ß gene (polß) in the mouse germ line results in a lethal phenotype. With the use of the bacteriophage-derived, site-specific recombinase Cre in a transgenic approach, the same mutation can be selectively introduced into a particular cellular compartment-in this case, T cells. The impact of the mutation on those cells can then be analyzed because the mutant animals are viable.
Gene targeting in embryonic stem (ES) cells provides a powerful tool for generating mice carrying predesigned mutations in the germ line (1). Current approaches to gene inactivation usually involve the introduction of a null mutation directly into ES cells from which homozygous mutant mice can be generated. Because the null mutation is carried in the germ line of the mutant animals, it will exert its effects from the onset of animal development. Although this approach to gene inactivation is valuable, for many applications it is important that the inactivation of a particular gene occurs in a conditional manner - for instance, in a predefined cell lineage or at a certain stage of development. Such conditional gene targeting would not only overcome problems posed by the fact that null mutations in the germ line are often lethal, but would also allow a more precise analysis of the impact of a mutation on individual cell lineages.
Somatic gene rearrangement and hypermutation at lymphocyte antigen receptor gene loci are unique events that require DNA repair (2, 3). The polß gene has been shown to be one of various enzymes involved in the DNA repair machinery (4). However, despite its ubiquitous expression (5) the importance of this enzyme for the generation of cell lineages, the survival of cells, and animal development in general remains elusive. To explore the function of the polß gene in mice, particularly in the development of lymphocytes, one attractive approach is to generate polß-deficient mice with the use of gene targeting. Because of the potential problem of embryonic lethality caused by a null mutation of the polß gene, we developed a general method for conditional gene inactivation with the use of the polß gene as a model. This method includes the concomitant production of a conventional (nonconditional) deletion mutant.

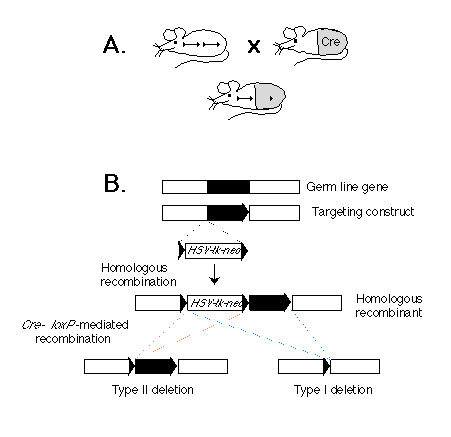
Fig. 1. (A) Scheme for cell type-specific gene targeting.
(Upper left) A mouse strain in which a gene in the germ line (black bar)
is flanked by two lox P sequences (triangles). (Upper right) A cre
transgenic mouse strain. The gray area represents the tissue in which the
Cre enzyme is expressed. (Bottom) F1 mouse derived from the two
mouse strains above. The target gene is deleted in the tissue in which the
Cre enzyme is expressed.
(B) Strategy for generating a gene flanked by lox P sites in ES cells.
Shown are a genetic locus, a corresponding targeting construct, the resulting
homologous recombinant, and the two types of Cre-mediated deletions. The
gene segment to be excised is depicted as a black bar. The lox P
sequences are represented by triangles. The position of the selection marker
cassette containing the neor and HSV-tk genes is indicated.
Our approach is based on the Cre-lox P recombination system of bacteriophage P1 (6). We and others have previously shown that this system is capable of mediating lox P site-specific recombination in both ES cells (7) and transgenic mice (8, 9). The strategy for conditional gene targeting is shown schematically in Fig. 1A. Two mouse strains are requited: One is a conventional transgenic strain in which a cre transgene is expressed in a cell type-specific or developmentally stage-specific manner. The second strain carries the target gene flanked by two lox P sites. In offspring derived from an intercross between these strains carrying the cre transgene and a lox P-flanked ("floxed") target gene, Cre-lox P site-dependent recombination will occur in cells where the cre gene is expressed, thereby deleting the target gene. In contrast, the target gene should remain functional in cells of all the other tissues, where the cre transgene is not expressed.
Depicted in Fig. 1B is a two-step strategy for generating in parallel a floxed gene or gene segment and a deletion of the same piece of DNA in ES cells in vitro. In the first step, three lox P sites, in addition to the selection marker genes for neomycin resistance (neor) and herpes simplex virus-thymidine kinase (HSV-tk), are introduced into the flanking regions of the target gene through homologous recombination. In the second step, the Cre enzyme is expressed in the genetically modified ES cells. If the expression of the Cre enzyme is transient, we expect that in some ES cells the recombination event will occur only once between any two of the three lox P sites, and different types of deletion should be generated. Type I deletion results in the deletion of the target gene from the genome of the ES cells, and animals derived from the corresponding ES cells will carry the deletion in the germ line. In contrast, type II deletion results in a floxed gene or gene segment at the targeted locus. The third possible type of deletion, which deletes the target gene but leaves the neor and HSV-tk genes in the genome, should not be observed in the mutant progeny, because ES cells carrying such a deletion should die after gancyclovir treatment during selection of type I and II deletion mutants (1).
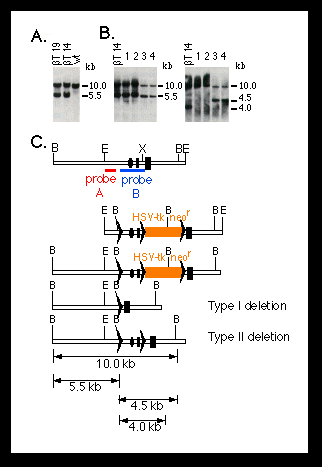
Fig. 2. Southern blot analysis of homologous recombination
and Cre-lox P-mediated recombination at the mouse polß locus.
The targeting and deletion experiments were performed as described (10)
(Fig. 1). DNA was digested with BamH I.
(A) Targeting of lox P sites into the polß locus of ES cells.
Shown are DNA samples from two candidate mutant ES cell clones (ßT14
and ßT19). The DNA from wild-type (wt) ES cells is shown as a control.
The 5.5-kb band represents the targeted polß allele.
(B) Cre-lox P-mediated deletion of the promoter and first exon of
the polß gene in ES cells. Genomic DNA was obtained from the parental
mutant ES cell (ßT14) and four subclones (1 to 4) carrying deletions
at the polß locus. Clones 1 and 2 carry a type I deletion, as shown
by a 5.5-kb band when they were hybridized with probe A (left panel) and
the absence of a 4.5-kb band when they were hybridized with probe B (right
panel) (C). Clones 3 and 4 have a type II deletion, as shown by a 4.5-kb
band when they were probed with probe B (C).
(C) Restriction maps for (from top to bottom) the 5' portion of the mouse
polß locus, the targeting construct, the homologous recombinant, and
the type I and type II deletion mutants. The dark rectangles represent the
first and second exons of the polß gene; the ovals represent the pole
promoter; the lox P sites are represented by triangles; and the red
and blue bars represent the probes used for hybridization. Restriction sites
of BamH I (B), EcoR I (E), and Xho I (X) are indicated. Numbers on the right
side of the blots indicate the sizes of the bands.
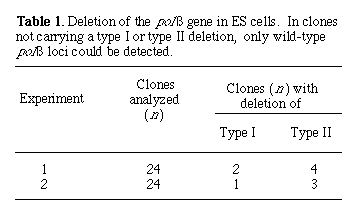
To generate polß mutant mice, we transfected the linearized targeting vector pMGß9 (Fig. 2C) into ES cells, and homologous recombinants were identified by Southern (DNA) blot analysis (10). Out of 288 G418-resistant clones analyzed, 16 were homologous recombinants, representing a frequency of 1 in 18. All these recombinants also carried a co-integrated lox P site approximately 1.5 kb upstream from the polß gene promoter (Fig. 2A). To generate type I and type II deletions, we transfected two mutant ES cell clones transiently with Cre-encoding plasmid DNA. Subclones carrying desired deletions at the polß locus were identified by Southern blot hybridization (Fig. 2B). In two independent experiments, both type I and type II deletions were consistently obtained (Table 1). For convenience, we refer to the type I deletion at the polß locus as polßD and the type Il deletion as polßflox.
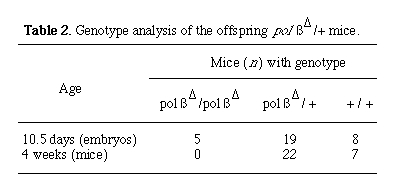
Mice carrying the polßD and polßflox mutations in the germ line were generated by the standard protocol (11). The impact of the polßD mutation was examined in offspring derived from an intercross between polßD heterozygous mice. No lethality was observed among the offspring from the time of birth to the age of 4 months. Genotypic examination of 4-week-old offspring revealed the absence of homozygous mutant animals (Table 2). However, at day 10.5 of fetal life, embryos homozygous for the polßD mutation were present at the frequency predicted by Mendelian laws (Table 2). On the basis of these results, we conclude that the homozygous mutant animals die in the course of fetal development.

As expected, animals homozygous for the polßflox mutation are viable. The overall development of these mutant mice also appears normal. These results indicate that the polßflox mutation does not severely hamper polß expression in vivo. Cell type-specific deletion of the polß gene was investigated in polßflox/+ mice carrying a cre lck transgene. The cre transgene in these mice is driven by the lck proximal promoter and is, therefore, selectively expressed in T lineage cells (9). The extent of polß gene deletion was assessed in various tissues of the animals by Southern hybridization (12). As expected, deletion occurred selectively in T cells. Quantitative analysis indicated that 63 to 84% of splenic T cells carried the deletion (Table 3). In contrast, no detectable deletion of the polß gene was observed in either kidney, liver, or B lymphocytes (Fig. 3A). These results demonstrate that cell type-specific gene inactivation can be achieved through our approach.
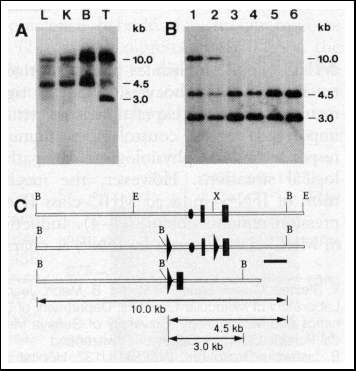
Fig. 3. Southern blot analysis of cell type-specific deletion
of the polß gene. DNA was obtained from various tissues of mutant
mice. T lineage and B cells were purified with a FACS sorter (12). Symbols
are as in Fig. 2.
(A) Cell type specificity of polß gene deletion. Shown are Southern
blot analyses of various tissues from polßflox/+; cre
lck transgenic mice. Genomic DNA was obtained from liver (L),
kidney (K), splenic B lymphocytes (B), and splenic Thy-1+ (T)
cells and digested with BamH I. The probe used for hybridization is indicated
in (C) as a black bar.
(B) Efficiency of polß gene deletion in T lineage cells. Samples were
obtained from mice of the following genotypes: polßflox/+;
cre lck (lanes 1 and 2), polßflox/polßD; cre lck (lane 3, 4, and 5), and polßflox/polßflox;
cre lck (lane 6). Lanes 1 and 3 represent DNA from thymic
CD4+ CD8+ cells; lanes 2, 4, and 6 from splenic Thy-1+
cells; and lane 5 from splenic B cells.
(C) Restriction maps of the 5' portion of the polß gene. Symbols are
as in Fig. 2.
To obtain T cells homozygous for the polßD mutation, we mated mice carrying the polßflox mutation and the cre lck transgene to heterozygous polßD mice. We chose for further analysis offspring of genotype polßflox/polßD that carried the cre lck transgene, because in such animals every single cell has only a single functional polß (namely, polßflox) gene, the deletion of which will result in homozygosity for the polßD mutation.
The overall development of polßD/ polßflox; cre lck transgenic mice appeared normal. Of these mice, both males and females were able to generate offspring when mated to normal mice, which suggests that germ cells developed normally in these animals. Flow cytometric analysis of T lineage cells in the thymus revealed no difference between the mutant mice and the wild-type controls in terms of total number of thymocytes and the distribution of CD4 and CD8 expression (13) on the surface of these cells (Fig. 4). In the blood of the mutants, essentially all T cells [as identified by the Thy-1 surface marker (13)] express the a/ ß T cell receptor (TCR), and the number of splenic Thy-1+ cells is also normal compared to that in wild-type mice (14).
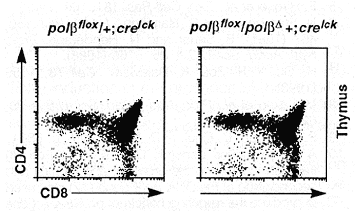
Fig. 4. Flow cytometric analysis of the thymocytes from mutant mice. The cells were stained with CD4 and CD8 antibodies and analyzed with a FACStar ( 12). The genotypes of the mice are indicated on top of each profile.
To estimate the fraction of T cells homozygous for the polßD mutation in these mice, we performed Southern blot analysis using DNA from purified T and B cells (Fig. 3B) (12). We found that approximately 40% of CD4+CD8+ thymocytes were homozygous for the polßD mutation (Table 3). The percentage of such cells was not increased in the peripheral T cells, which suggests that no further deletion of the polß gene occurred in T cells after the CD4+ CD8+ stage of T cell development in the thymus. This is consistent with the observation that the cre lck transgene is not expressed in mature T cells (9).
There may be two main reasons for the incompleteness of polß gene deletion in our experimental system. First, the lck proximal promoter is active only at early stages of T cell development (15). Earlier data also indicate that the cre lck transgene is expressed only transiently in the thymus (9). It is therefore conceivable that in the transgenic T cells the polßflox alleles have only a brief period of time to accomplish Cre-lox P-mediated recombination. Second, the cre gene that we have used corresponds to the wild-type cre gene of P1 phage (6). It is known that the expression of this gene in eukaryotic cells is suboptimal, but it can be improved by appropriate genetic manipulation (7, 16). Thus, there are straightforward ways in which our experimental system can be improved to obtain a more efficient deletion of the target gene.
Our data provide no direct evidence at this stage about a possible involvement of polß in the control of TCR gene rearrangements. However, we might interpret the lesser extent of polß deletion in polßD/ polßflox; cre lck mice as compared to that in polßflox/+; cre lck transgenic mice (Table 3) to mean that in the former case, Cre-lox P-mediated polß inactivation results in cell death if it happens to occur before the completion of TCR gene rearrangement.
In principle, Cre-lox P-mediated gene targeting should allow the inactivation of any gene in any tissue at any stage of development. It can also be adapted to conditional reconstitution of gene function. Furthermore, through lineage-specific inactivation of genes critical for cell survival, this approach can potentially be used for the ablation of cell lineages in vivo.
REFERENCES AND NOTES
1. K. R. Thomas and M. R. Capecchi, Cell 51, 503 (1987) .
2. F. W. Alt, T. K. Blackwell, G. D. Yancopoulos, Science 238, 1079 (1987).
3. S. Brenner and C. Milstein, Nature 21 1, 242 (1966); C. Kocks and K. Rajewsky, Anna. Rev Immunol. 7, 537 (1989).
4. M. Fry, in Enymes of Nucleic Acid Synthesis and Modification, S. T. Jacob, Ed. (CRC Press, Boca Raton, FL, 1983), pp. 39 92.
5. F. Hirose et al., Exp. Cell Res. 181, 169 (1989).
6. N. Sternberg and D. Hamilton, J. Mol. Biol. 150, 467 (1981); B. Sauer and N. Henderson, Prod. Natl. Acad Sci. U.S.A. 85, 5166 (1988).
7. H. Gu, Y.-R. Zou, K. Rajewsky, Cell 73, 1155 (1993).
8. M. Lakso et at, Pros. Natl. Acad Sci. U. S.A. 89, 6232 (1992).
9. P. C. Orban, D. Chui, J. D. Marth, ibid., p. 6861.
10. A 7.1-kb mouse genomic DNA (EcoR I-EcoR I) fragment containing the promoter and the first and second exon of the DNA polß gene (17) was used to produce the targeting construct pMGß9. A gene cassette containing the neor and HSV-tk genes finked by two lox P sites was inserted into the Xho I site between the first and second exon. A third lox P site was introduced into a Sac I site approximately 2 kb upstream from the first exon. The final targeting construct contains 1 kb of flanking genomic sequences further upstream from the third lox P site and a 3-kb fragment including the second exon of the polß gene downstream from the Xho I site (Fig. 2C). To generate homologous recombinants, we transfected E14-1 ES cells [R. Kuhn, K. Rajewsky, W. Moller, Scence 254, 707 (1991)] with 25 µg of DNA (of the linearized targeting construct) by electroporation. The transfected ES cells were grown on a single layer of mitomycin C-treated embryonic fibroblasts. After 1 week of selection in G418-containing medium, homologous recombinants were identified by Southern blot hybridization based on the strategy depicted in Fig. 2C. A targeted clone should yield a 5.5-kb band in addition to an equally intense 10-kb wild-type band upon hybridization to probe A (Fig. 2). To generate type I and type II deletions, 1 to 3 µg of supercoiled Cre-encoding plasmid, pIC-Cre (6), was introduced into the targeted ES cells by electroporation. After selection in ganciclovir-containing medium (1 x 10-6 M) for 5 days, surviving clones were picked and expanded. Genomic DNA was then prepared from the expanded cells for Southem blot analysis. Probe A is a 1-kb genomic DNA fragment (Hind III-Sac I) of the polß gene, and probe B is a 900-bp fragment (BamH I-Hind III) (17).
11. A. Bradley, in Teratocarcinomas and Embronic Stem Cell: A Practical Approach, E. J. Robertson Ed. (IRL Press, Oxford, 1987), pp. 113 157.
12. On the basis of the expression of specific cell surface markers, we purified T and B cells by fluorescence-activated cell sorting using a FACStar (Becton Dickinson). T cells were sorted as Thy-1+ ceils from the spleen. The CD4+ CD8+ cells were from the thymus. B cells were sorted from the spleen as surface CD45R/B220+ cells. Usually, 5 x 106 to 1 x 107 sorted cells were used for DNA preparation. The Southern hybridization was performed according to the standard protocol [J. Sambrook, E. F. Fritsch, T. Maniatis, Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1989)] with DNA from 2 x 106 sorted cells. The probe was a 900-bp BamH I-Hind III fragment of the polß gene (17). Densitometric analysis was performed with a Bio-Imaging analyzer (Fuji) or a densitometric scanner (Pharmacia).
13. For a detailed description of thymic T cell development, see J. Sprent [in Fundamental Immunology, W. E. Paul Ed. (Raven Press, New York, 1993), pp. 75-109] .
14. H. Gu and K. Rajewsky, unpublished data.
15. O. Sartor et al., Mol. Cell. Biol. 9, 2983 (1989); A. M. Garvin et al., Int. Immunol. 2,1973 (1990); K. E. Chaffin etat, EMBO J. 9, 3821 (1990).
16. B. Sauer and N. Henderson, New Bio. 2, 441 (1990).
17. M. Yamaguchi et al., Mol. Cell Biol. 7, 2012 (1987) .
18. We thank A. Matsukage for providing the plasmid containing the genomic pole gene and R. Kuhn for the Et4-1 ES cell line. We are grateful to R. Torres, F. Huetz, and Y.-R. Zou for critical reading of the manuscript; to U. Ringeisen for graphical work; to W. Moller for help at many levels; and to all our colleagues for helpful discussion. Supported by the Deutsche Forschungsgemeinschaft through SFB 243, the Fazit Foundation, the Bundesministerium for Forschung und Technologie, and the Human Frontier Science Program.
Return To Molecular Biology Main Page
Return To Biology Dept. Main Page
© Copyright
2000 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu