Nature VOL. 337 12 JANUARY,
1989
The bicoid protein is a positive regulator of
hunchback transcription in the
early Drosophila embryo
Wolfgang Driever & Christiane Nüsslein-Volhard
Max Planck Institut für Entwicklungsbiologie, Abteilung 111
Genetik, Spemannstrasse 35, 7400 Tübingen, FRG
A gradient in concentration of the protein product of the bicoid
gene is a determinant of the anterior-posterior axis of Drosophila
embryos. By binding upstream of the segmentation gene hunchback
the bicoid protein controls its transcription, thereby translating
maternal pattern-generating information into differential activation
of zygotic gene expression.
THE molecular link between the maternal genome and the activation
of zygotic gene expression is realized by maternal factors deposited
in the egg during oogenesis. Three groups of maternal genes specify
distinct domains along the anteroposterior axis of the Drosophila
melanogaster embryo1. In
the anterior group the maternal gene bicoid (bcd
) is crucial for the development of head and thorax2. bcd mRNA is localized at the
anterior pole of the egg and early embryo3,4
and the bcd protein comes to be distributed in a concentration
gradient with a maximum at the anterior tip of the embryo5. The bcd protein gradient seems to determine
position in the anterior half of the embryo in a concentration-dependent
way6. The presence of a homoeodomain7 in the bcd protein suggests that it
binds to DNA in a sequence-specific way8,9
and thereby regulates the spatially restricted expression of zygotic
target genes. The earliest zygotic genes expressed in distinct
spatial domains along the anterior-posterior axis of the embryo
are the gap genes10. The phenotype
of mutations in the gap gene hunchback (hb; deletion
of gnathal and thorasic segments11),
and the expression pattern of hb RNA in bcd mutant
embryos12 identify hb as a probable
target for bcd regulation. Here, we show that the bcd protein
binds to five sites upstream of the transcription start site of
the zygotic gap gene hunchback These binding sites define
the consensus binding sequence: 5' TCTAATCCC 3'. Transient expression
assays in embryos as well as in tissue culture cells reveal that
the three binding sites in the promoter region (-50 to -300) are
necessary and sufficient for the activation of zygotic hunchback
expression.
Mapping bcd protein binding sites
In the syncytial blastoderm hb expression occurs in two
domains (ref. 13, see also Fig. 3a). The 2.9-kilobase (kb) transcript
in the anterior domain is under the control of a separate promoter
and is not expressed in bcd mutant embryos12.
The regulatory region necessary for correct spatial expression
of the zygotic 2.9 kb hb transcript has been mapped using p-transformation
of hb ß-galactosidase gene fusions14
to a 700-base pair (bp) DNA fragment that includes the first intron
and 300 bp upstream of the transcription start site.
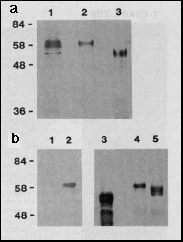 Fig. 1 a, Expression of bcd full-length
protein in E. coli and Drosophila Schneider cell
tissue culture. Western blot of extracts separated by PAGE, probed
with anti-bcd antibodies. Lane 1, extracts from 2- to 3-hour-old
Drosophila melanogaster embryos (500 µg protein);
lane 2, extracts from the Schneider cell line S2/M3 that transiently
expresses bcd under the control of the Drosophila actin
5c promoter (plasmid pPAcbcdEE, 100 µg protein applied);
lane 3, extracts from E. coli that express bcd under the
control of the T7 phi 10 promoter (plasmid pARbcdNB, 10 µg
protein applied); Mrs are indicated
on the left, in thousands.
Fig. 1 a, Expression of bcd full-length
protein in E. coli and Drosophila Schneider cell
tissue culture. Western blot of extracts separated by PAGE, probed
with anti-bcd antibodies. Lane 1, extracts from 2- to 3-hour-old
Drosophila melanogaster embryos (500 µg protein);
lane 2, extracts from the Schneider cell line S2/M3 that transiently
expresses bcd under the control of the Drosophila actin
5c promoter (plasmid pPAcbcdEE, 100 µg protein applied);
lane 3, extracts from E. coli that express bcd under the
control of the T7 phi 10 promoter (plasmid pARbcdNB, 10 µg
protein applied); Mrs are indicated
on the left, in thousands.
b, Modification of bcd protein by phosphorylation. The bcd protein
was transiently expressed in Schneider cells grown in the presence
of [32P]orthophosphate. Immunoprecipitation
from nuclear extracts with anti-bcd antibody and autoradiography
of the precipitate resolved on SDS-PAGE demonstrates that in bcd-expressing
cells (lane 2) but not in non-expressing cells (lane 1) a phosphorylated
form of bcd protein is synthesized. Incubating bcd immunoprecipitates
from extracts of Schneider cells transiently expressing bcd protein
with potato acid phosphatase results in an increase of the electrophoretic
mobility of bcd protein. The most rapidly migrating form is of
the same size as the bcd protein expressed in E. coli (lane
3) as revealed from western blot analysis of the reaction products:
bcd protein incubated without (lane 4), and with (lane 5) potato
acid phosphatase.
Methods. The bicoid gene gives rise to a primary
transcript that is subject to differential splicing. The alternatively
spliced RNAs code for different protein products3,5.
In this study we used only one protein species derived from cDNA
c53.46.6 (ref. 3). Although it is not yet clear whether the different
forms of bcd protein are functionally equivalent, RNA injections
into early embryos reveal that this bcd protein species can rescue
all aspects of the mutant bicoid phenotype (our unpublished
data). For the protein expression constructs, bcd cDNA
c53.46.6c was used (ref. 3; all the base pairs that differ from
the genomic sequence and give rise to amino-acid substitutions
were exchanged with genomic sequences: detailed protocol on request).
Construction of pARbcdNB: the EcoRI-SalI fragment from c53.46.6c
was used to generate a NdeI site at the initiator ATG of the longest
open reading frame by oligonucleotide site-directed mutagenesis
(using the pMa/c 254 plasmid vectors, Stanssens and co-workers,
unpublished results), the cDNA was reconstructed and a BamHI site
generated at the XmaI site 3' to the open reading frame; the NdeI-BamHI
fragment was cloned into the expression vector pAR3038 (ref. 15).
Construction of pAcbcdEE: the EcoRI fragment of cDNA c53.46.6c
was blunt-end-ligated into the BamHI site of the vector pPAc downstream
of the actin 5c promoter and upstream of the actin 5c polyadenylation
processing signals (vector pPAc constructed by H. Lipshitz using
the Drosophila melanogaster actin 5c gene, ref. 28). bcd
protein was expressed from pARbcdNB (ref. 15) in E. coli BL
21 DE3. Drosophila tissue culture and transfection techniques
were as described29. Phosphate
labeling was performed using 0.5 mCi 32P-labeled
NaH2PO4
per 5 ml of medium in a 6 cm petri dish. bcd protein was immunoprecipitated
from cleared lysates of isolated nuclei sonicated in 20 mM Tris
pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1 mM dithiothreitol, 10% glycerol,
0.5% NP-40, 2 mM phenylmethylsulphonylfluoride using polyclonal
anti-bcd antibody prebound to Staph. aureus (Calbiochem).
The immunoprecipitates where treated with 50 µg ml-1 DNaseI (Cooper), 10 mM MgCl2in the above buffer for 15 min on ice.
Potato acid phosphatase treatment (20 µg per 100 µl)
was as described30.. Extract
preparation from Drosophila embryos, electrophoresis and
further processing were as previously described5..
Full-length bcd protein was expressed in Escherichia coli
by generating a NdeI site at the start ATG of the longest open
reading frame and cloning bcd-protein-coding sequences into the
pAR3038 expression vector15.
The same cDNA was also expressed in Drosophila Schneider
cells using the actin 5c promoter (Fig. l a). The protein derived
from E. coli migrates slightly faster (lane 3, apparent
relative molecular mass Mr=
53,000 (53K)) than that from Schneider cells (lane 2, 58K) or
embryos (lane 1, 56-60K). Different electrophoretic mobilities
can be explained by post-translational modification. To test whether
the decrease in electrophoretic mobility is due to phosphorylation
(see for example engrailed (en)16.
Ultrabithorax (Ubx), L. Gavis, personal communication),
we immunoprecipitated bcd protein transiently expressed in Schneider
cells which had been grown on [32P]
orthophosphate-containing media. The immunoprecipitate was analyzed
by SDS-PAGE. A 32P-labeled protein
migrated at the expected size of the bcd protein (Fig. 1 b, lane
2). Treatment of immunoprecipitates from Schneider cells transiently
expressing bcd protein with potato acid phosphatase results in
a decrease in apparent Mr of
the immunoprecipitated protein (lane 4, 5), which now co-migrates
with the bcd protein expressed in E. coli (lane 3). In
addition, multiple bands of intermediate apparent Mr are also visible. We conclude that the
bcd protein in Drosophila Schneider cells, and probably
also in the embryo, is subject to multiple phosphorylations.
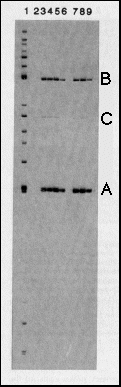 Fig. 2 Specific binding of bcd protein
to restriction fragments from the hb gene. The genomic
7.5 kb hb BamHI-EcoRI fragment contains the zygotic 2.9 kb transcript
that is expressed in the anterior gap domain and about 4.5 kb
of upstream sequences12. It
was digested with a set of restriction enzymes to produce fragments
of 100-1,000 bp and the fragments were end-labeled (lane 1). bcd
protein expressed in E. coli under the control of T7 RNA
polymerase (see Fig. 1), was used to immunoprecipitate fragments
that bind specifically to the protein. Lane 2 shows a control
reaction using anti-bcd immunoprecipitates from non-bcd expressing
bacteria; no fragments were bound. In the immunoprecipitates with
bcd protein (lanes 3-9), two fragments (A, a 243-bp SalI-MluI
fragment from -298 to -55 with respect to the transcription start
site; B. a 459-bp HindIII-BglII fragment from -2,172 to -2,631)
were bound with high affinity, one with relatively low affinity
(C) a BamHI-HindIII fragment from -4,242 to -3,890). To assess
the specificity of binding, zero (lane 3), 0.25 (lane 4), 1 (lane
5) and 5 (lane-6) µg of competitor DNA (salmon sperm DNA,
1,000-fold excess over total labeled DNA, 40,000-fold excess over
one specific labeled fragment, lane 6) were included in the binding
reaction. Binding of fragments A and B is only slightly affected
at the highest competitor concentration, fragment C is no longer
bound under this condition. In the experiments shown in lanes
7, 8 and 9, after 45 min binding the immunoprecipitates were washed
and competitor DNA was added for 10 min (0.2, 1 and 5 µg,
respectively). The off rates for binding to fragment A and B are
apparently rather low.
Fig. 2 Specific binding of bcd protein
to restriction fragments from the hb gene. The genomic
7.5 kb hb BamHI-EcoRI fragment contains the zygotic 2.9 kb transcript
that is expressed in the anterior gap domain and about 4.5 kb
of upstream sequences12. It
was digested with a set of restriction enzymes to produce fragments
of 100-1,000 bp and the fragments were end-labeled (lane 1). bcd
protein expressed in E. coli under the control of T7 RNA
polymerase (see Fig. 1), was used to immunoprecipitate fragments
that bind specifically to the protein. Lane 2 shows a control
reaction using anti-bcd immunoprecipitates from non-bcd expressing
bacteria; no fragments were bound. In the immunoprecipitates with
bcd protein (lanes 3-9), two fragments (A, a 243-bp SalI-MluI
fragment from -298 to -55 with respect to the transcription start
site; B. a 459-bp HindIII-BglII fragment from -2,172 to -2,631)
were bound with high affinity, one with relatively low affinity
(C) a BamHI-HindIII fragment from -4,242 to -3,890). To assess
the specificity of binding, zero (lane 3), 0.25 (lane 4), 1 (lane
5) and 5 (lane-6) µg of competitor DNA (salmon sperm DNA,
1,000-fold excess over total labeled DNA, 40,000-fold excess over
one specific labeled fragment, lane 6) were included in the binding
reaction. Binding of fragments A and B is only slightly affected
at the highest competitor concentration, fragment C is no longer
bound under this condition. In the experiments shown in lanes
7, 8 and 9, after 45 min binding the immunoprecipitates were washed
and competitor DNA was added for 10 min (0.2, 1 and 5 µg,
respectively). The off rates for binding to fragment A and B are
apparently rather low.
Methods. The genomic 7.5 kb BamHI-EcoRI fragment was isolated
from the plasmid pE8-B1000A (obtained from D. Tautz, ref. 13),
digested with a set of restriction enzymes and end-labeled with
polynucleotide kinase (restriction enzymes: AvaII, BglII, MluI,
HindIII, NdeI, SalI and NcoI; no additional fragments were precipitated
when the same experiments were performed using either a HinfI
or a TaqI digest). bcd protein was expressed in the bacterial
strain BL21(DE3) as described in ref. 15. Induced cells were collected
by centrifugation and resuspended in 1:200 volume of buffer Z
(ref. 21), incubated for 15 min on ice and sonicated twice for
15 s. The lysate was spun clear and the supernatant taken as extract.
Immunoprecipitation was carried out according to a modified procedures.
Fixed Staph. aureus cells (Calbiochem, 100 µl of
10% suspension) were washed twice with binding buffer BB (20 mM
Tris pH 7.5, 50 mM NaCl, 0.5 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol,
10% glycerol) and incubated with polyclonal anti-bcd antibody
(20 µg in 500 µl BB) for one hour on ice. The suspension
was washed twice with BB. Extracts (100 µl) from bacteria
carrying pARbcdNB or pAR3038, respectively, were added in 1 ml
BB, 2 mM PMSF and incubated for two hours on ice. The immunocomplexes
were washed twice with BB, once with BB 0.2% NP-40 and resuspended
in 100 µl BB. Binding reactions were in 50 µl total
volume including 10 µl immunocomplex suspension, NaCl added
to 170 mM, 5 ng end-labeled fragments and various amounts of sonicated
salmon sperm DNA as competitor for 40 min on ice. For lanes 7-9,
immunoprecipitates were washed once with BB and incubated for
10 min in the above reaction mixture with the indicated amounts
of competitor DNA. Precipitates were washed four times with I
ml BB 0.2% NP-40 each, resuspended in 100 µl 10 mM Tris
pH 7.5, 1 mM EDTA, 5 µg sonicated salmon sperm DNA was added
as carrier, the fragments phenol/chloroform extracted, precipitated
and separated on a 4% denaturing polyacrylamide gel. Autoradiography
was performed using Cronex 4 films.
We used an immunoprecipitation assay17
to screen large regions of genomic hb DNA for bcd-binding sites
(see Fig. 2). E. coli -derived full-length bcd protein
was immunoprecipitated with polyclonal antibodies bound to fixed
Staphylococcus aureus cells and the resulting immunoprecipitate
was incubated with end-labeled restriction fragments from the
hb promoter. Two restriction fragments from the hb
gene were bound with high affinity: fragment A (-298 to -55) and
B (-2,172 to -2,631) base pairs upstream from the transcription
start site of the zygotic 2.9 kb hb transcript (Fig. 3a).
A third fragment, C, from -4,242 to -3,890 base pairs upstream
from the 2.9 kb transcription start site (or -1,020 to -672 upstream
from the 3.2 kb transcript) was a bound with relatively low affinity.
Fragment A binds with the highest affinity as, even at the highest
concentrations used, salmon sperm DNA had a negligible effect
on the amount of fragment A bound. At this competitor concentration
the binding of fragment B is reduced to about one-third and binding
of fragment C is eliminated completely. The same fragments were
also precipitated using bcd protein expressed in Schneider cells
(data not shown), indicating that phosphorylation does not affect
binding specificity in this assay.
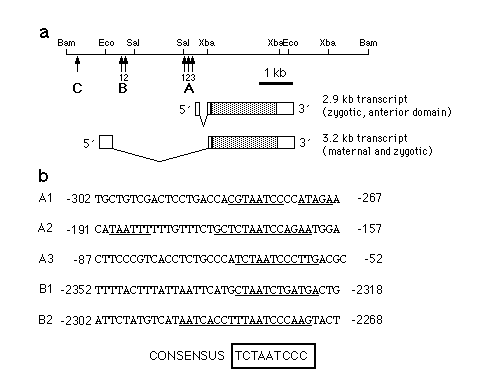
Fig. 3 a, Map of the hb gene indicating the locations
of bcd-binding sites. During the syncytial blastoderm stage hb
expression occurs in two domains which are under separate regulation.
The 2.9 kb hb transcript is expressed in an anterior domain
which extends from 55-100% egg length, whereas the 3.2 kb transcript
(which is expressed maternally and zygotically) is localized to
0-25% egg length. In addition, the 3.2 kb transcript can be detected
in a narrow stripe at about 53% egg length shortly before the
onset of cellularization. A, B and C are the fragments identified
in the experiment shown in Fig. 2. b, Nucleotide sequences of
the regions where bcd protein binds to hb regulatory regions.
Base pairs protected against DNaseI digestion as referred from
Fig. 4 are indicated by a bar below the sequence. Comparison of
the protected sequence elements led to the identification of a
consensus binding sequence. Bases in the protected regions that
fit the consensus sequence are in bold.
Consensus sequence for bcd binding
The individual binding sites were analyzed further by DNaseI footprint
experiments13. The results are
shown in Fig. 4 and summarized in Fig. 3. Fragment A contains
three regions which are protected against DNaseI digestion by
bcd protein, Al (-280 bp), A2 (-170 bp) and A3 (-65 bp). A2 consists
of two protected regions, one well protected (around -170) and
one weakly protected (around -187). The A region is part of the
700 bp fragment shown to be sufficient for the correct spatial
expression of the 2.9 kb hb transcript14.
The B fragment consists, of two binding regions, B1 (about -2,325)
and B2 (about -2,280). Both are less well protected than the A
sites. No footprint could be obtained with the C fragment (data
not shown), but DNaseI hypersensitive sites could be identified
around -4,100 and -3,998. Analysis of the sequences of fragment
A and B revealed that each of the sites contains a sequence approximating
a 9-bp consensus sequence TCTAATCCC (Fig. 3b). This sequence is
best conserved in sites A2 and A3 whereas A1, Bl and B2 each contain
mismatches with the consensus. Only the central part of the sequence,
TAATC is conserved in all five sites. Using binding conditions
of low stringency (no competitor DNA) hypersensitive sites could
be detected around CTAAT or TAATC sequences (for example -4,100,
-3,998) or even at TAAT alone (data not shown). In addition, within
fragment A in the region -241 to -204 on the sense strand some
hypersensitive sites do appear and one can detect weak protection
of some bases on the antisense strand. Three sequence motifs in
this region (TGCTAAGCT, -241 to -233; (GCTAAGCT,
-225 to -217; and GATCATCC, -212 to -205) resemble
the consensus sequence (bold nucleotides) weakly. Notably, all
three motifs are palindromic. The bcd protein seems to bind to
DNA sequences that contain elements like TAAT, CTAA, CTAAT, TAATC,
where the context of the surrounding nucleotides determines the
affinity of binding, the best fit so far identified being the
consensus TCTAATCCC.
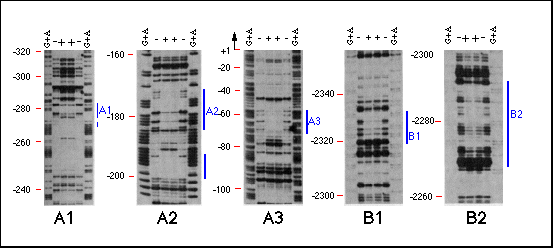
Fig. 4 Analysis of bcd protein binding to hb
sequences by DNaseI footprinting. The DNA fragments identified
in the immunoprecipitation assay, the sequences surrounding these
fragments as well as the intron of the 2.9-kb transcript were
analyzed for bcd protein binding sites by DNaseI footprinting.
Specific binding could only be detected in the same regions that
were identified by the immunoprecipitation assay regions A and
B. Region A seemed to consist of three bcd binding sites, Al,
A2 and A3, each spaced at about 100-bp intervals. Region B consists
of two binding sites, about 40 bp distant from one another. Lanes
show (G+A) sequence marker and footprint reactions with (+) and
without ( - ) bcd protein. Positions are indicated on the left
side of each panel in base pairs distance from the transcription
start site. Protected regions are marked by a bar on the right
side.
Methods. For DNaseI protection assays (ref. 31) bcd protein
was affinity purified as described in Fig. 2 and resuspended in
100 µl FPBB21 (110 mM
KCl, 47 mM HEPES pH 7.8, 12 mM MgCl2,
0.05 mM EDTA, 1 mM dithiothreitol, 17% glycerol and 0.02% NP 40).
Genomic fragments were asymmetrically labeled with Klenow and
radioactive dNTPs (Al: labeled at MluI site -55, linearized at
NsiI site -434, sense strand; A2 and A3: labeled at an EcoRI site
generated at SalI-298 and linearized at XbaI+431, antisense strand,
B1 and B2 labeled at an EcoRI site generated at the BglII site
-2,170 linearized at HindIII -2,631, sense strand). Binding reactions
were done with 4-8 ng of labeled fragment and 20 µl of immunocomplex
in a total volume of 50 µl FPBB for 45 min on ice. 50 µl
10 mM MgCl2/ 5 mM CaCl2 were added to the reaction followed
by 5 µl of freshly diluted DNaseI (Worthington) at a final
concentration of 5 µg per ml reaction mix. After 5 min on
ice, digestion was stopped by the addition of 110 µl 1%
SDS, 20 mM EDTA, 200 mM KCl, 250 µg per ml yeast tRNA (65°
C). After phenol / chloroform extraction and ethanol precipitation
samples were electrophoresed on 8% polyacrylamide / 7.5 M urea
gels and visualized by autoradiography.
For a few other Drosophila homoeodomain-containing proteins,
sequence-specific binding has been demonstrated and the binding
sites seem to lack palindromic character as does the consensus
sequence. Engrailed (en), fushi tarazu (ftz),
zerknüllt (zen) and even skipped (eve)
protein in vitro all bind en upstream sequences
with the consensus TCAATTAAT20,21.
In addition, eve protein recognizes TCAGCACCG sequences in the
eve upstream region21.
Ubx protein binds to (TAA)n
repeats (P. Beachy, M. Krasnow, L. Gavis and D. Hogness, personal
communications). Analyzing the "contact amino acids"
of the homeodomain that are probably exposed to DNA (positions
1, 2, 5, 6 and 9 within the recognition helix of the helix-turn-helix
motif22, one finds that Ubx,
ftz, zen and en are identical in all but
the second amino acid, providing an explanation for their similar
binding characteristics. In contrast, bcd differs in positions
1, 2 and 9 with respect to the above proteins. Further analysis
will show if bcd has overlapping binding characteristics
with other Drosophila homeodomain proteins.
Embryonic expression assay
To investigate the influence of the bcd-binding sites on hb transcription,
we used an embryonic transient expression assay (Fig. 5). The
hb promoter fragments, including increasing numbers of
bcd-binding sites, were fused to the chloramphenicol acetyltransferase
(CAT) gene at position +107 of the 2.9 kb hb transcript. The plasmid
constructs were injected into the anterior half of early cleavage
embryos and the expression of the reporter gene CAT was measured
at the onset of gastrulation. We expect that a small amount of
the plasmid DNA ends up in a nuclear location during the rapid
early cleavage cycles and comes to be expressed under conditions
similar to those that lead to the expression of the chromosomal
hb copies.
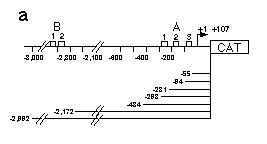
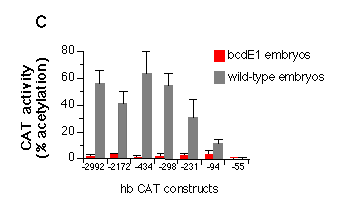
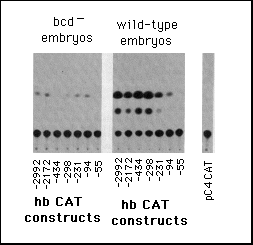
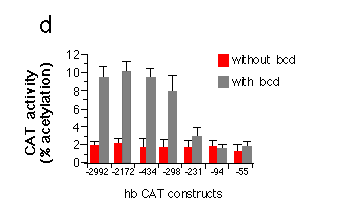
Fig. 5 Influence of hb upstream sequences on the expression
of a reporter gene coding for CAT as assayed by transient expression
in embryos and Schneider cell tissue culture.
a, Map of hb CAT fusion genes used in this analysis. hb
upstream sequences and the hb transcription start site
plus 107 bp of the non-translated leader were fused to the CAT
gene. Construct hbCAT-55 ends just downstream of the A3 binding
site, hbCAT-94 includes the A3 binding site, hbCAT-231 includes
A2 and A3, hbCAT-298 the whole A cluster. To analyze a possible
function of the B cluster, we made constructs ending upstream
and downstream of that cluster.
b, Embryonic transient expression assay. hbCAT fusion gene plasmids
were injected into early cleavage embryos and the embryos analyzed
for expression of CAT at the onset of gastrulation. The left part
shows CAT assays using embryos from homozygous bcdE1
females (control with no bcd protein) and the right part those
from wild-type embryos. pC4 CAT marks the control when just injecting
the vector alone without any hb sequences.
c, Quantitative analysis of the embryonic transient expression
assay. Percentage of acetylated forms of total chloramphenicol
were plotted for the expression of CAT in wild-type and bcdE1 embryos. The stimulation of expression
in wild-type embryos compared to bcd mutant embryos is about 50-fold
when all three sites of the A cluster are present, 10-fold with
just A2 and A3 and 3-fold when only A3 is present. Comparing the
construct that has no bcd-binding site (hbCAT-55) with the one
that contains all three A sites (hbCAT -298) gives a 200-fold
stimulation of expression. The presence or absence of the B cluster
does not seem to have a significant influence on the expression
starting from the zygotic 2.9 kb hb transcript start site.
d, Transient expression of hb-CAT fusion genes in Drosophila
Schneider cells co-transfected with bcd under the control
of the actin 5c promoter (pPAcbcdEE, see Fig. 1). Correction for
variation in transfection efficiency was achieved by co-transfection
of an actin 5c LacZ fusion gene and assay of ß-galactosidase
activity in the cell extracts. The graph shows activation of CAT
expression when comparing transfections with or without bcd.
All hb-CAT constructs showed a high background expression in the
absence of bcd. The presence of bcd protein leads to a
fivefold stimulation of CAT expression from constructs containing
all three A sites. Removal of Al results in a strong reduction
of expression. CAT expression in the presence of just A3 is no
more different from background levels. Northern analysis of RNA
from cells transfected with similar hb lacZ fusion genes
indicate an about fourfold increase in transcript level comparing
the hb-55lacZ to the hb -298lacZ construct.
Methods. A BamHI site was generated at the TaqI site within
the zygotic 2.9 kb hb transcript (+107) by ligating a BamHI linker
to the filled &laqno; in TaqI site. Genomic hb DNA was digested
with enzymes cutting at the sites indicated in a, blunt-ended
and then cut at the newly introduced | BamHI site (+107). The
CAT reporter gene vector pC4 CAT (ref. 32) was linearized at the
SalI site in the polylinker, blunt-ended and cut i with BamHI.
Isolated hb promoter fragments were cloned into the pC4 CAT vector
by standard techniques. Each plasmid (5 pmol) dissolved in 0.1
ml sterile water containing 106 c.p.m. 32P dCTP. Injections into
early cleavage embryos, 100 of each mounted on one cover and covered
with Voltalef 1OS oil, were according to standard procedures2.
The embryos were left to develop at 18° C until the onset
of gastrulation, washed off the coverslip with heptane, washed
carefully three times with heptane to remove last traces of oil
and the heptane was evaporated completely with an air stream.
Radioactivity injected into the embryos was determined and the
embryos were frozen in liquid nitrogen. An average of 70 µl
was injected into each 100-embryo batch (about 7% of the egg volume).
Batches with injected amounts more than 30% different from the
average were discarded. For each wild-type construct four groups
of 100 embryos and for the bcdE1 control as(d > well as the
vector control two groups of 100 embryos were analyzed. Modified
CAT assays were used33. 100 µl of 0.25 M Tris pH 7 8 was
added to each batch, the embryos sonified for 5 s, incubated at
65 šC for 5 min and insoluble material discarded after centrifugation.
Aliquots (25 >1) of the extract were mixed with 54 µl
0.25 M Tris pH 7.8, 20 >1 4 mM acetylCoA and I µl [14C]chloramphenicol
(0.2 ,µCi). The reaction mixture was incubated for one hour
at 37° C, the chloramphenicol extracted with ethyl acetate
(0.5 ml), the ethyl acetate evaporated and the acetylated forms
of chloramphenicol separated by TLC. The acetylated and non-acetylated
forms of chloramphenicol were visualized by autoradiography and
quantitated by scintillation counting. Bars in c show mean values
of the determinations and the standard error of the I mean. Drosophila
tissue culture and transfection techniques were used29. The
pPAcLacZ plasmid was constructed by inserting the blunt ended
| SalI-HindIII fragment from pCaSpeR AUG GAL (ref. 32) containing
the coding region for ß-galactosidase and the SV40 polyadenylation
site into a blunt-ended SalI-digested pPAc actin 5c promoter vector
(H. Lipshitz, unpublished results). Co-transfections were performed
using 5 µg pPAcLacZ, equimolar amounts of the hb-CAT constructs
(1-1.5 µg) with or without 10 µg of pPAcbcd. pUC 18
DNA was added to reach 20 µg total DNA per transfection.
Cells from one 6-cm plate were washed in PBS and Iysed in 200
µl 250 mM Tris pH 7.8 by sonication. Cleared Iysates were
assayed for ß-galactosidase activity using o-nitrophenyl-,B
D galactopyranoside as substrate. The measured activities were
used to correct the amount of extract used in the CAT assay (see
above) for the variation in transfection efficiency. The graph
shows mean values of two determinations and the mean errors.
On injection of the hb-CAT constructs into early embryos, we
do indeed observe a bcd-dependent expression of CAT. Figure 5
shows that with the longest upstream sequences CAT expression
is about 50 times higher in embryos from wild-type females than
in embryos derived from bcd- females. Elimination of the binding
sites B1 and B2 did not significantly affect the level of expression.
In contrast the successive elimination of the A sites results
in a dramatic reduction of the bcd-dependent hb-CAT expression.
If all binding sites (but not the TATA box) are removed, no significant
expression is observed.
We also expressed the CAT constructs in Schneider cells. In
this assay, there is a substantial expression of CAT even in the
absence of bcd. Co-transfection (M. Krasnow and D. Hogness, unpublished
results) with the bcd transcription unit under the control of
the actin 5c promoter results in a fivefold stimulation of CAT
expression using constructs that contain the whole A region (Fig.
5d ). The strong reduction in expression when removing only the
Al site supports the idea that the three sites cooperatively activate
hb expression. Deletion of A1 and A2 decreases the expression
to background levels.
Discussion
The concerted action of three regulatory sites, each 100 bp apart,
raises the question of how they interact with the distant |tanscription
initiation complex. In prokaryotes, loop formation between promoter
elements has been visualized on a molecular level23.
In eukaryotes, the chromatin structure might influence the spatial
arrangement of regulatory regions. The analysis of the Drosophila
26K heat-shock protein promoter24
demonstrates that the positioning of a nucleosome might bring
regulatory sequences together on its surface25.
The spacing of bcd-binding sites A1-A3 might allow a similar mechanism.
Remarkably, in Drosophila virilis, not only the bcd-binding
sites, but also their spacing seem to be conserved (D. Tautz,
personal communication). A close spatial proximity of the binding
sites might be the basis for cooperativity in activation of transcription.
An interesting question is whether bcd protein provides only the
DNA-binding function or also the transcription-activating functions
that interact with the cellular transcription machinery. In yeast,
transcriptional activators are characterized by acidic protein
domains26 and these acidic regions
seem to function in other eukaryotic systems as well27. A good candidate for a transcriptional
activator domain within the bcd protein is the region between
amino acids 345 and 390 (see ref. 3 for protein sequence) that
has 10 negative charges and in addition several putative phosphorylation
sites. As noted above, the protein is phosphorylated in Drosophila
Schneider cells. The level of phosphorylation might influence
the strength of the transcriptional activator function of bcd.
The molecular interaction between the bcd protein and the hb promoter
is probably one of the first events in gene regulation during
embryogenesis and shows how a maternally derived regulatory protein,
bcd, participates in initiating the process of pattern formation.
A major question about the interactions between maternal coordinate
genes and zygotic segmentation genes is how the information provided
by the smoothly graded distribution of a maternal gene product
like bcd is transformed into the more sharply defined gap gene
domains. In the wild-type, the early posterior border of hb expression
is at 55% egg length12 . In
this region of the embryo the bcd gradient is already very shallow,
although the bcd protein is detectable up to 30% egg length5. Several models might explain how the
transition from a shallow gradient to a sharp on/off decision
is achieved. For example, other so far unidentified factors might
compete for bcd-binding sites or inhibit bcd binding and thereby
sharpen the posterior border of hb expression. So far there is
no genetic evidence for the existence of genes that could function
as competitors1, 2, 6. Our data
support models that include cooperativity as an important mechanism
to control the spatially restricted expression of zygotic target
genes. A precedent for the role of cooperativity in gene regulation
is the action of l repressor (for review
see ref. 19). The three bcd-binding sites identified in the hb
promoter might allow cooperative binding and thereby full activation
of transcription above a low threshold of bcd concentration. Indeed,
the level of hb expression is constant within the anterior 40%
of the embryo12, independently
of the marked increase in bcd protein concentration towards more
anterior positions. In addition, a cooperative mechanism can generate
an on/off transition within a relatively small range of bcd protein
concentrations. One would not expect that such a mechanism immediately
generates a sharp border; indeed, initially the posterior border
of hb expression is not clearly defined within a stripe two or
three nuclei wide12. Subsequent
regulatory interactions between hb and other zygotic genes might
be involved in the establishment and maintenance of a border as
sharp as that observed in later blastoderm stages12.
Genetic analysis suggests that bcd regulates more than one zygotic
target gene in a spatially restricted way26.
Different spatial limits of target gene expression could be achieved
if the affinities of their bcd-binding sites vary. Thus a reduced
affinity for bcd protein binding would restrict target gene expression
to more anterior regions which contain higher levels of bcd protein.
Analysis of further target genes will show how bcd protein fulfills
its function as a morphogen at the molecular level.
We thank G. Thoma for technical assistance, P. O'Farrell, J. Ma,
M. Ptashne, T. Hoey, L. Gavis, M. Krasnow and J. Adamczewski for
discussion, P. Stanssens, H. J. Fritz (pMa/c), H. Lipshitz, C.
S. Thummel, A. M. Boulet (pPAc, pC4CAT, pCaSpeR AUGbGal)
for providing plasmid vectors J. Pohlner for oligonucleotide synthesis,
and our colleagues D. St Johnston, S. Cohen, V. Siegel, H. and
R. Schnabel, M. Frasch, L. Stevens, M. Klingler and S. Roth for
critically reading the manuscript. Photographs were prepared by
R. Groemke Lutz. The work was supported by the DFG (Leibniz programme).
Received 6 October; accepted 22 November 1988.
1. Nüsslein-Volhard, C., Frohnhöfer, H. G. & Lehmann,
R. Science 238,1675-1681 (1987).
2. Frohnhöfer, H. G. & Nüsslein-Volhard, C. Nature
324, 120-125 (1986).
3. Berleth, T. et al. EMBO J. 7, 1749-1756 (1988).
4. Frigerio, G., Burri, M., Bopp, D., Baumganner, S. & Noll,
M. Cell 47, 735-746 (1986).
5. Driever, W. & Nüsslein-Volhard, C. Cell 54,
83-93 (1988).
6. Driever, W. & Nüsslein-Volhard, C. Cell 54,
95 104 (1988).
7. McGinnis. W., Levine, M. S., Hafen, E., Kuroiwa, A. & Gehring,
W. J. Nature 308, 428-433 (1984).
8. Laughon, A. & Scott, M. P. Nature 310, 25-31
(1984).
9. Desplan, C., Thies, J. & O'Farrell, P. H. Nature
318, 630-635 (1985).
10. Knipple, D. C., Seifert, E., Rosenberg, U. B., Preiss, A.
& Jäckle, H. Nature 317, 40-44 (1986).
11. Lehmann, R. & Nusslein-Volhard, C. Dev. Biol. 119,
402-417 (1987).
12. Tautz, D. et al. Nature 332, 281-284 (1988).
13. Tautz, D. et al. Nature 327, 383-389 (1987).
14 Schröder, C. et al. EMBO J. 17, 2881-2888
(1988).
15. Studier, F. W. & Moflat, B. A. J. Molec. Biol.
189, 113-130 (1986).
16. Gay, N. J., Poole, S. J. & Kornberg, T. H. Nucleic
Acids Res. 16, 6637-6647 (1988).
17. McKay, R. J. Molec. Biol. 145, 471-488 (1981).
18. Galas, D. & Schmitz, A. Nucleic Acids Res. 5,
3157-3170 (1978).
19. Ptashne, M. A Genetic Switch (Cell Press, Palo Afto/Blackwell,
Oxford, 1986).
20. Desplan, C., Theis, J. & O'FarreB, P. Cell 54,
1081-1090 (1988).
21. Hoey, T. & Levine, M. Nature 332, 858-861
(1988).
22. Pabo, C. O. & Sauer, R. T. A. Rev. Biochem. 53,
293-321 (1984).
23. Griffith, J., Hochschild, A. & Ptashne, M. Nature
322, 750-752 (1986).
24. Thomas, G. M. & Elgin, S. C. R. EMBO J. 7,
2191-2201 (1988).
25. Weintraub, H. Nucleic Acids Res. 8, 4745-4753
(1980).
26. Ma, J. & Ptashne, M. Cell 48, 847-853 (1987).
27. Kakidani, H. & Ptashne, M. Cell 52, 161-167
(1988).
28. Bond, B. J. & Davidson, N. Molec. Cell Biol. 6,
2080-2988 (1986).
29. Rio, D. C. & Rubin, G. M. Molec. Cell Biol. 5,
1833-1838 (1985).
30. Cooper, J. A. & King, C. S. Molec. Cell Biol. 6,
4467-4477 (1986).
31. Heberlein, U., England, B. & Tjian, R. Cell 41,
956-977 (1985).
32. Thummel, C. S., Boulet, A. M. & Lipshitz, H. D. Gene
(in the press).
33. Gorman, C. M., Moffat, L. F. & Howard, B. H. Molec.
Cell Biol. 2, 1044-1051 (1982).
Return
To Molecular Biology Main Page
Return To Course
Materials

©
Copyright 2000 Department of Biology, Davidson College, Davidson,
NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu