| Jump to: | |
The genome of baker's yeast, Saccharomyces cerevisiae, has been fully sequenced but not fully annotated. On this page, we will explore the extent of knowledge about an annotated gene, MBP1. We will then search the data for information about a non-annotated gene, YDL057W, in order to make an educated hypothesis as to its function. The following diagram shows the location of both the annotated MBP1 and the non-annotated YDL057W on the IV chromosome of S. cerevisiae:
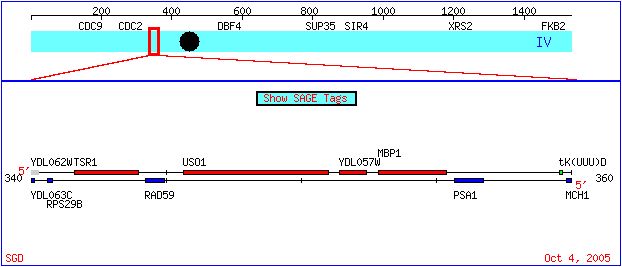
Figure 1. The location of YDL057W and MBP1 on the S. cerevisiae chromosome IV. Diagram taken from the Saccharomyces cerevisiae Genome Database.
MBP1 is short for MluI-box Binding Protein, and it codes for a transcription factor. The nucleotide sequence (from the S. cerevisiae Genome Database) of MBP1 is shown here. The location of the gene on the chromosome is shown below (Figure 2).

Figure 2. This figure shows the location of MBP1 on the S. cerevisiae chromosome IV. From the NCBI MapViewer.
The following figure shows the BLASTn results for MBP1.
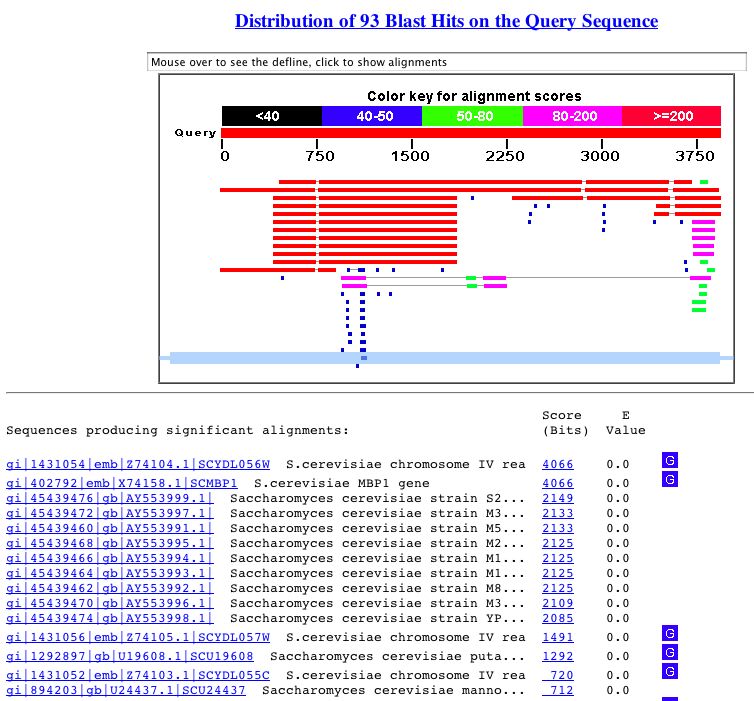
Figure 3. This shows the results of a BLASTn search with the nucleotide sequence of MBP1. As you can see, the genes that are significantly similar (as shown by their low E-value) are simply instances of the gene in other strains of S. cerevisiae.
There are several conserved domains found on the MBP1 gene, as shown by the conserved domain search below:
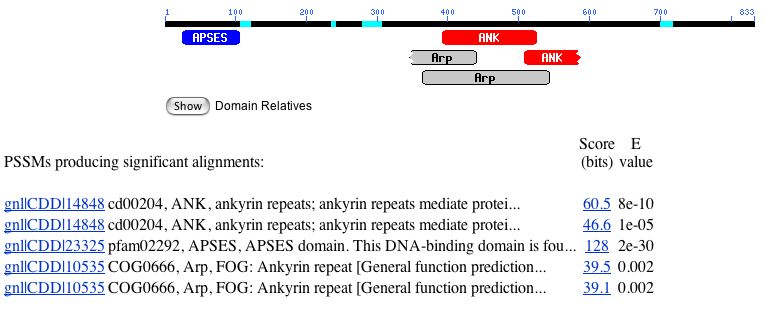
Figure 4. A conserved domain search for MBP1 yielded five conserved domains.
As aforementioned, MBP1 encodes a transcription factor. The transcription factor is 833 amino acids long and has a molecular weight of 93,907 Da. The following is the amino acid sequence of the MBP1 protein:
MSNQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRIL EKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDFTQTDGS ASPPPAPKHHHASKVDRKKAIRSASTSAIMETKRNNKKAEENQFQSSKILGNPTA APRKRGRPVGSTRGSRRKLGVNLQRSQSDMGFPRPAIPNSSISTTQLPSIRSTMGP QSPTLGILEEERHDSRQQQPQQNNSAQFKEIDLEDGLSSDVEPSQQLQQVFNQNT GFVPQQQSSLIQTQQTESMATSVSSSPSLPTSPGDFADSNPFEERFPGGGTSPIISMI PRYPVTSRPQTSDINDKVNKYLSKLVDYFISNEMKSNKSLPQVLLHPPPHSAPYID APIDPELHTAFHWACSMGNLPIAEALYEAGTSIRSTNSQGQTPLMRSSLFHNSYT RRTFPRIFQLLHETVFDIDSQSQTVIHHIVKRKSTTPSAVYYLDVVLSKIKDFSPQY RIELLLNTQDKNGDTALHIASKNGDVVFFNTLVKMGALTTISNKEGLTANEIMNQ QYEQMMIQNGTNQHVNSSNTDLNIHVNTNNIETKNDVNSMVIMSPVSPSDYITY PSQIATNISRNIPNVVNSMKQMASIYNDLHEQHDNEIKSLQKTLKSISKTKIQVSL KTLEVLKESSKDENGEAQTNDDFEILSRLQEQNTKKLRKRLIRYKRLIKQKLEYR QTVLLNKLIEDETQATTNNTVEKDNNTLERLELAQELTMLQLQRKNKLSSLVKK FEDNAKIHKYRRIIREGTEMNIEEVDSSLDVILQTLIANNNKNKGAEQIITISNANS HA |
Completing a Kyte-Doolittle analysis to determine the protein's structure yields the following graph:
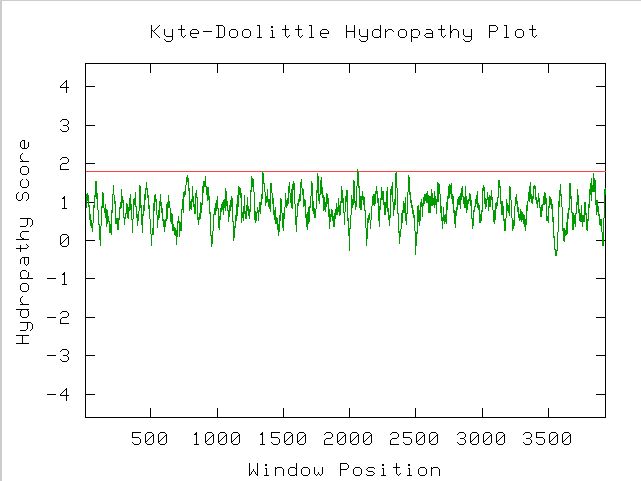
Figure 5. A Kyte-Doolittle Hydropathy Plot used to predict the structure of a protein. Because the peaks of the graph reach over a 1.8 hydropathy score at least once, there is most likely a transmembrane region on the MBP1 protein.
Another method of predicting the structure of a protein is shown below:
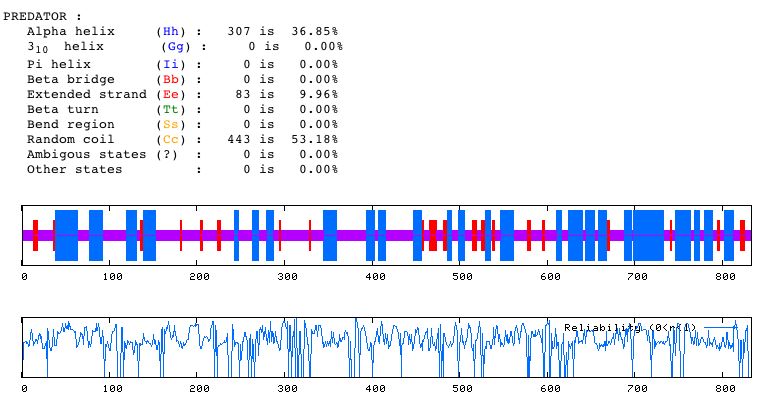
Figure 6. The Predator Secondary Structure Prediction Method was used to determine that the MBP1 protein secondary structure consists mostly of random coils, while around 37% of the characterizable morphological regions of the protein are alpha helixes.
The following diagram is the result of a BLASTp search to find amino acid sequences similar to that of the MBP1 transcription factor.
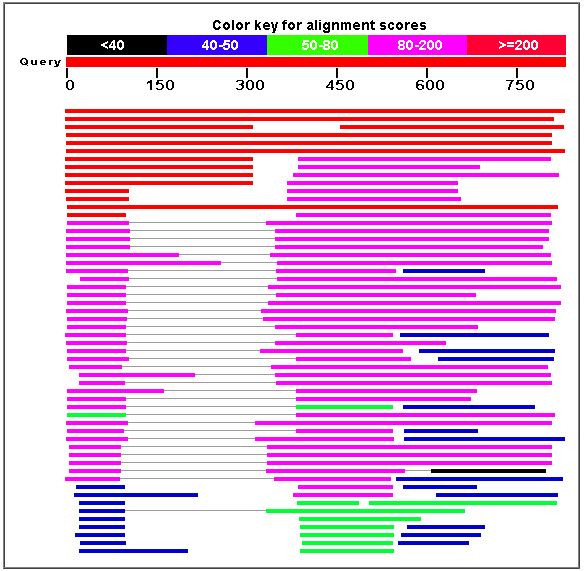
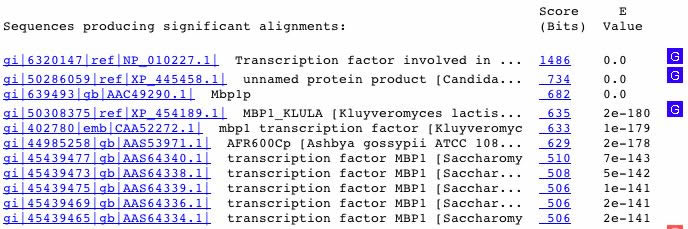
Figure 7. A BLASTp search yielded proteins similar to MBP1. The similar proteins included transcription factors in other species, such as Kluyveromyces lactis and Candida glabrata.
The protein function, according to NCBI Gene, is regulation of the cell cycle progression from the G1 phase to the S phase. The following screenshot from NCBI Gene details the protein's molecular function, biological process, and cellular component:
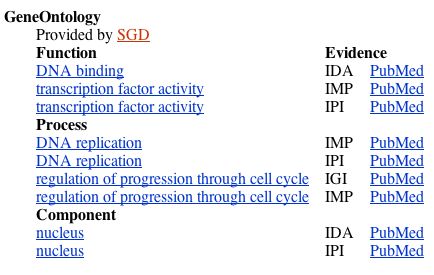
Figure 8. The molecular function, biological process, and cellular component of the MBP1 transcription factor, from NCBI Gene.
The nucleotide sequence for the non-annotated gene YDL057W is available here. The following diagram shows the position of YDL057W on the S. cerevisiae chromosome IV:

Figure 9. The position of YDL057W on S. cerevisiae chromosome IV. From the NCBI MapViewer.
A BLASTn search with the sequence of YDL057W yielded the following results:
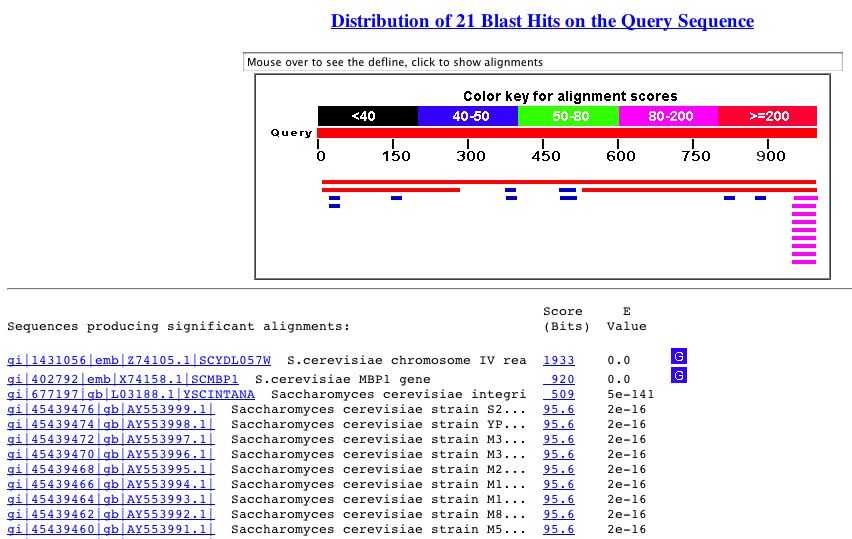
Figure 9. The BLASTn results from a query with the nucleotide sequence of YDL057W. As you can see, there is significant similarity between YDL057W and an integrin-like gene in S. cerevisiae.
The following conserved domains were found on the YDL057W gene:

Figure 10. Conserved domains for YDL057W, from the Conserved Domain Database.
The protein coded for by YDL057W has the following amino acid sequence:
MEKKHVTVQIQSAPPSYIKLEANEKFVYITSTMNGLSYQIAAIVSYPEKRNSSTAN KEDGKLLCKENKLALLLHGSQSHKNAIYQTLLAKRLAEFGYWVLRIDFRGQGDS SDNCDPGLGRTLAQDLEDLSTVYQTVSDRSLRVQLYKTSTISLDVVVAHSRGSL AMFKFCLKLHAAESPLPSHLINCAGRYDGRGLIERCTRLHPHWQAEGGFWANGP RNGEYKDFWIPLSETYSIAGVCVPEFATIPQTCSVMSCYGMCDHIVPISAASNYAR LFEGRHSLKLIENADHNYYGIEGDPNALGLPIRRGRVNYSPLVVDLIMEYLQDT
|
A BLASTp search produced the following results:
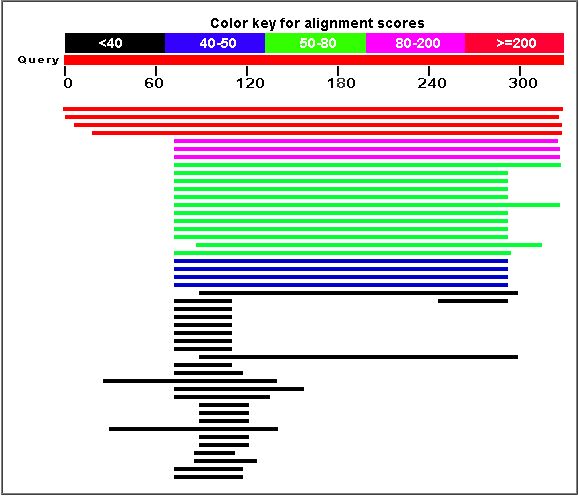

The following are orthologs of the YDL057W gene:
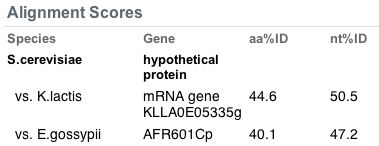
Figure 12. Orthologs of the YDL057W gene. The highest similarity found was in K. lactis with 50% nucleotide similarity.
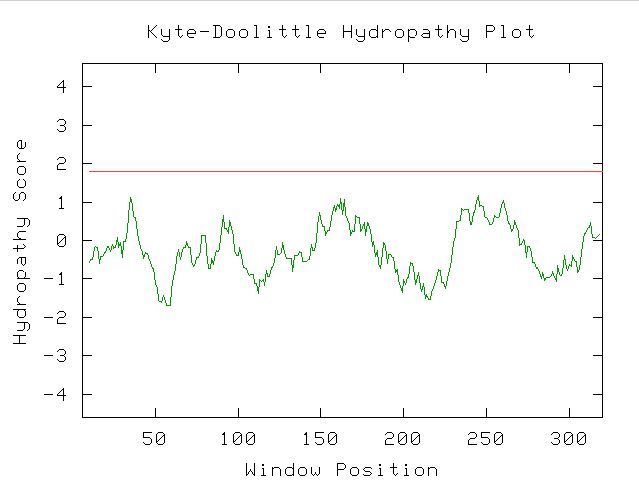
Figure 13. Kyte-Doolittle Hydropathy Plot showing that there are no transmembrane regions in YDL057W, since the hydropathy score never exceeds 1.8.
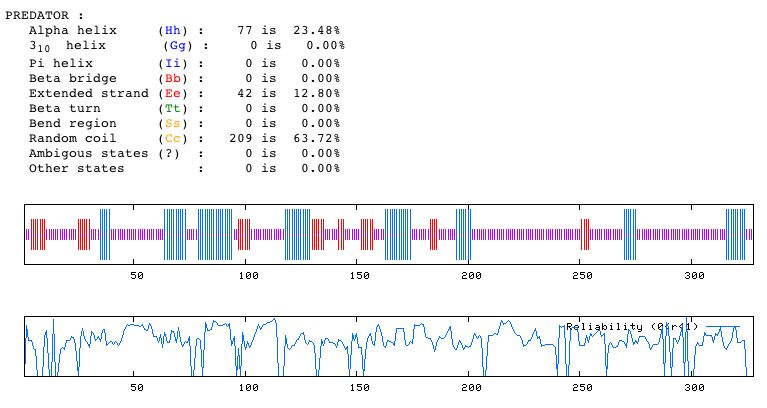
Figure 14. Predator analysis predicting the secondary structure of YDL057W. Note that most of the characterizable morphological details of the secondary structure are random coils.
Because the BLASTn analysis showed that YDL057W was very similar to an integrin-like gene in S. cerevisiae, my first instinct was that YDL057W also coded for an integrin-like protein. There is some speculation that an integrin-like gene in baker's yeast could be a prototype for the later integrin-like proteins in eukaryotes. However, integrins are cell adhesion and cell signalling molecules, and therefore it is difficult for me to believe that an integrin-like protein would lack a transmembrane region. Thus, it is difficult to hypothesize about the protein for which YDL057W may code. I do think that a cell adhesion molecule, perhaps one that has no signalling function (and therefore does not need a transmembrane region), is the best guest based on the BLASTn search.
Hodge, C.W., Mehmert, K.K., Kelley, S.P., McMahon, T., Haywood, A., Olive, M.F., Wang, D., Sanchez-Perez, A.M., and R.O. Messing. (1999) Supersensitivity to allosteric GABA(A) receptor modulators and alcohol in mice lacking PKCepsilon. Nat. Neurosci. 2(11): 997-1002.
Author Last Name, Initials. Date page created or revised. Title of page. Title of larger work if applicable. <URL>. Accession date.
Kyte-Doolittle Hydropathy Plot. 2005. <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm > Accessed 2005 7 Oct.
NCBI. 2005. National Center for Biotechnology Information. <http://www.ncbi.nih.gov/> Accessed 2005 7 Oct.
PREDATOR Database 2005. <http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html> Accessed 2005 7 Oct.
Saccharomyces Genome Database. 2005. <http://www.yeastgenome.org/> Accessed 2005 7 Oct.