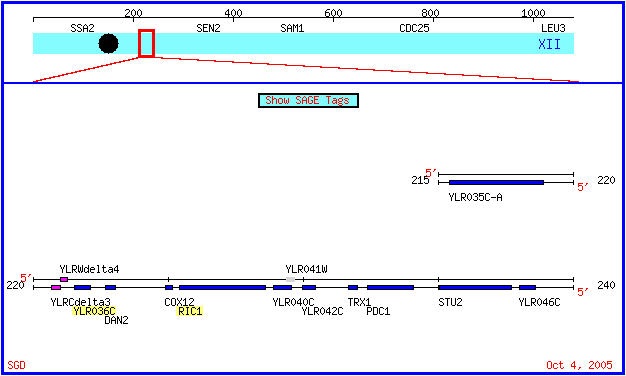
Figure 1: A map of chromosome 12 in Saccharomyces cerevisiae. The Ric1 and YLR036 genes are highlighted. Both genes are on the Crick strand. (SGD, 2005; http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000004029).
Annotated Yeast Gene: Ric1
Sequence
The genomic, coding and amino acid sequences of ric1 are available at the Saccharomyces Genome Database.
Gene Ontology
The molecular function and cellular component of Ric1 are both known. Ric1 has been identified as a peripheral membrane protein specific to the Golgi apparatus (Siniossoglou et al., 2000). It functions in the fusion of the Golgi apparatus with vesicles derived from endosomes. The molecular function of Ric1 is dependent upon multiple other proteins, including Rgp1p. The Ric1 and Rgp1p proteins form a heterodimeric complex that activates Ypt6p, a GTPase that is also localizes to the exterior of the Golgi membrane. The activation occurs through a guanine nucleotide exchange on the Ypt6p (Siniossoglou et al., 2000).
Ric1 is known to function in at least two biological processes involving the membrane of the Golgi apparatus. Through its activation of the Ypt6p GTPase, Ric1 functions in the fusion between the Golgi and vesicles derived from endosomes. Ypt6p interacts with SNARE proteins to coordinate membrane fusion between the Golgi and vesicles. Once activated, Ypt6p triggers a series of interactions that assist in SNARE binding, which ultimately induces fusion of the Golgi membrane with vesicles (Siniossoglou et al., 2001). Studies also indicate that Ric1 functions in the localization of the trans-Golgi network of membrane proteins (Bensen et al., 2001).
Mutation Studies
Although viable, ric1 mutants fail to properly transcribe ribosomal protein genes and ribosomal RNA (SGD, 2003; http://db.yeastgenome.org/cgi-bin/phenotype/phenotype.pl?dbid=S000004029). Also, a genome-wide examination of haploinsufficiency found that ric1 mutants have reduced fitness in rich medium (Deutschbauer et al., 2005). Deletion of ric1 resulted in the mislocalization of two trans-Golgi network proteins, Kex2p and Vps10p, which supports Ric1's proposed role in TGN localization (Bensen et al., 2001).
Hydropathy Profile
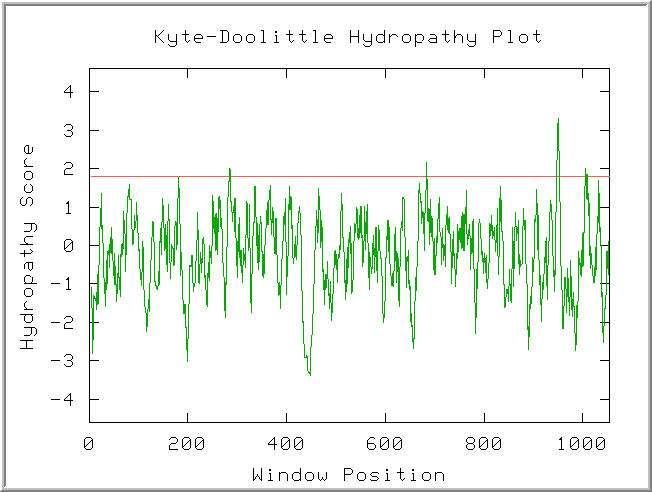
Figure 2: Kyte-Doolitle Hydropathy plot of the Ric1 polypeptide. Default settings. (KDHP, 2002; http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm).
Four peaks in the plot were found to have hydropathy scores above 1.8. The hydropathy score agrees with the proposed model for Rip1 as a peripheral membrane protein. The four hydropathy maxima might funciton to anchor Rip1 to the Golgi membrane.
Blastp Search
The Blastp service at NCBI.gov was used to search for proteins with sequences similar to Ric1. The results are shown in Figure 3.
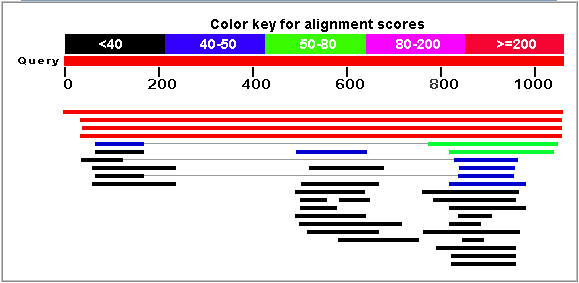
Figure 3: Blastp results for the Rip1 protein. The top hit (red line) corresponds to the Ric1 protein sequence. (NCBI, 2005; http://www.ncbi.nlm.nih.gov/BLAST/)
The two strongest matches were unnamed protein products found in the two fungi Candida glabrata and Kluyveromyces lactis. The strongest non-fungi match was found in a protein from Tetrahymena thermophila (E = 0.28). Also, Ric1 was matched with a hypothetical malarial antigen (E = 0.82). No specific functional trend was observed in the results.
Conserved Domains
A search for conserved domains within the Ric1 amino acid sequence found one match (Figure 4).

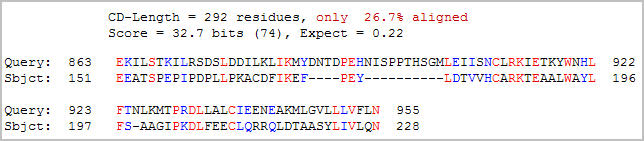
Figure 4: Conserved domain analysis of the Ric1 protein. A DUF1339 domain was aligned with Ric1 with an expect value of 0.22 (NCBI, 2005; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Although the expect value (0.22) is suggestive of a relationship, little information on the DUF1339 domain is available. The DUF13339 domain is a hypothetical eukaryotic protein that is possible an integral membrane protein. Interestingly, the DUF1339 domain is located in the same stretch of Ric1 that yielded the highest hydropathy scores in the Kyte-Doolittle plot. Since both proteins are believed to be bound to membranes, the observed hydropathy in the conserved region can be interpreted as evidence of conservation between the two proteins.
Non-Annotated Gene: YLR036C
Sequence Information
Genomic, coding and amino acid sequences are available at the Saccharomyces Genome Database.
Gene Ontology
Molecular Function: Unknown
Biological Process: Unknown
Cellular Component: Unknown
Mutation Studies
A genome-wide deletion study found that yeast strains with a mutant version of YLR036C are viable (Giaever G et al., 2002).
Blastn
A Blastn search returned no hits (NCBI, 2005, http://www.ncbi.nlm.nih.gov/BLAST/).
Protein Statistics
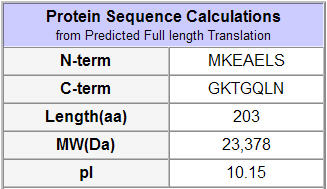
Figure 5: Protein statistics on the predicted translation of the YLR036C ORF (SGD, 2002; http://db.yeastgenome.org/cgi-bin/protein/protein?sgdid=S000004026).
SGD predicted some basic information on the protein product (above). The data describe a predicted translation that might differ from the actual protein product.
Hydropathy Profile
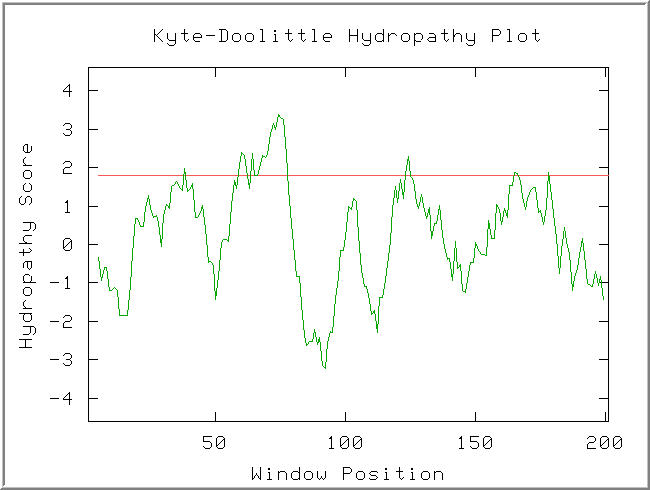
Figure 6: Kyte-Doolittle hydropathy plot of the YLR036c protein. Default Settings.(KDHP, 2002; http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm).
A strongly hydrophathic region is observed in the window at AA70. The width of this hydropathic sequence suggests it might be a transmembrane region.
Blastp
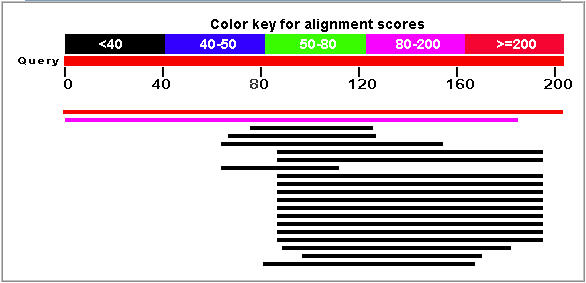
Figure 7: Blastp results for the YLR036c protein. The top hit (red line) corresponds to the YLR036c protein. (NCBI, 2005; http://www.ncbi.nlm.nih.gov/BLAST/).
The pink line in Figure 7 corresponds to a strong match with the hypothetical protein YIR089wp that is also found in Saccharomyces cerevisiae (E = 3e-20). The Blastp search did not find another protein sequence that gave an expect value of less than 0.1. The unparalleled similarity between these two yeast proteins is highly suggestive of a duplication event in Saccharomyces cerevisiae.
The YIR089w translation is an integral membrane protein. Since YLR036c contains an apparent transmembrane region (see Figure 6) and is highly similar to YIR089w, it is likely an integral membrane protein.
The numerous Blastp hits between amino acids 80 and 200 was interpreted as evidence of a conserved domain. The region appears to show greater conservation than other portions of the YLR036C gene. The conservation suggests that the region has evolved at a slower rate and is associated with the function of the YLR036c function.
Conserved Domains
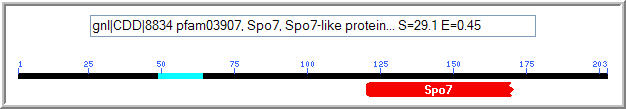

Figure 8: Conserved domain analysis of the YLR036c protein. A Spo7 domain was matched to YLR036c with an expect value of 0.45 (NCBI, 2005; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
The Spo7 gene encodes an integral membrane protein associated with either the nucleus or the endoplasmic reticulum. It is specific to Saccharomyces cerevisiae, and functions in meiosis and sporulation. (NCBI, 2005; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene&cmd=Retrieve&dopt=full_report&list_uids=851224). It is known to function in meiosis and sporulation. The Kyte-Doolittle plot of YLR036c (Figure 6) showed evidence of a transmembrane region, although it was not found in the portion of YLR036c that features conservation with the Spo7 protein.
Secondary Structure
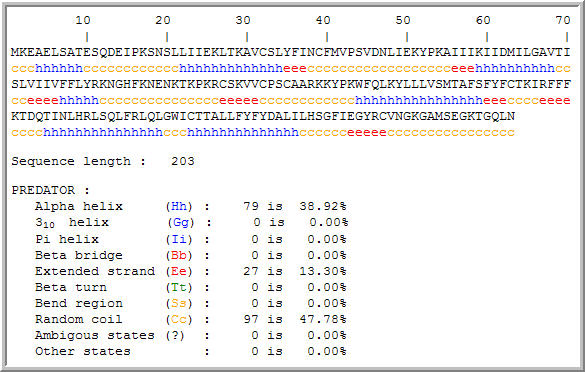
Figure 8: Secondary structure motifs in the hypothetical YLR036c protein. The secondary structure information might aid in further analysis of protein function. (PREDATOR, 2005; http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html)
Predicted Function
Considering the data, YLR036c appears to encode an integral membrane protein. A broad hydrophathic region was observed in the Kyte-Doolittle hydropathy plot of the YLR036c protein. Also, the YLR036c was aligned with the integral membrane protein YIR089w with an expect value of 3e-20.
The conservation between YLR036c and Spo7 is suggestive of a similar function. The region conserved between YLR036c and Spo7 appeared to be the most highly conserved region in the YLR036c protein based on the Blastp search. Spo7 is known to function in sporulation and meiosis, so I hypothesize that the YLR036c is involved in the same processes. Although sporulation and meiosis are essential functions in yeast, redundancy with the YIR089w protein might compensate for loss of the YLR036c protein in mutant strains.
References
1 Siniossoglou S, Peak-Chew, SY, Pelham HRB. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. J European Molecular Biology Organization 2000 July; 19(18): 4885-4894. p 4885.
2 Siniossoglou S, Pelham HRB. An effector of Ypt6p binds the SNARE Tig1p and mediates selective fusion of vesicles with late Golgi membranes. J European Molecular Biology Organization 2001 September; 20(21): 4885-4894. p 4885.
3 Bensen ES, Yeung BG, Payne GS. Ric1p and the Ypt6p GTPase Function in a Common Pathway Required for Localization of Trans-Golgi Network Membrane Proteins. J The American Society for Cell Biology 2001 January; 12: 13-26. p 13.
4 Deutschbauer AM, Jaramillo DE, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficency revealed by genome-wide profiling in yeast. Genetics 2005 April; 169(4): 1915-25. p 1915.
5 Giaever, G et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002 July; 418(6896):387-391.
Resources
Kyle-Doolittle Plot Form. 2002. < http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm> Accessed 2005 5 October.
NCBI. 2004. National Center for Biotechnology Information. <http://www.ncbi.nih.gov/> Accessed 2005 5 Oct.
PREDATOR Database 2004. <http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html> Accessed 2005 5 Oct.
Saccharomyces Genome Database. 2004. <http://www.yeastgenome.org/> Accessed 2005 5 Oct.
Ben Whigham's Genomics Home Site.
Genomics Course Web Site.
© Copyright 2005 Department of Biology, Davidson College,Davidson, NC 28036
Send comments, questions, and suggestions to: bewhigham@davidson.edu