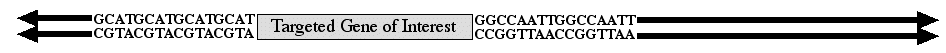
When an investigator wants to replace one allele with an engineered construct but not affect any other locus in the genome, then the method of choice is homologous recombination. To perform homolgous recombination, you must know the DNA sequence of the gene you want to replace (figure 1). With this information, it is possible to replace any gene with a DNA construct of your choosing. The method has a few more details than will be illustrated on this page, but the essential information is retained.
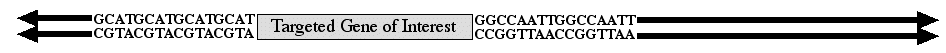
Figure 1. Diagram of gene targeted for replacement by an engineered construct. The coding sequence is illustrated by the box with flanking upstream and downstream DNA sequences provided. The arrows pointing away from the targeted gene represent the continuous chromosomal DNA.
The next step is to design and fabricate the DNA construct you want to insert into the chromosome in place of the wild-type allele. This construct may contain any DNA sequence of your choosing which means you can insert different alleles (both functional and non-functional ones), different genes or reporter genes (e.g. antibiotic resistance or green fluorescent protein). Regardless of what you want to insert, you must include some flanking DNA that is identical in sequence to the targeted locus (figure 2). In addition to the positive selection marker (e.g. antibiotic resistance) often a negative selection marker (e.g. thymidine kinase, tk) is added to the replacement vector. The negative marker is outside the region of sequence similarity between the vector and the targeted locus.

Fiugre 2. Diagram of engineered construct that will be used to replace the wild-type allele. The upstream and downstream flanking DNA sequences are identical to those which flank the targeted locus. The negative marker tk is shown in to the right of the region of sequence similarity.
The engineered construct is added to cells which contain the targeted gene of interest. By mechanisms that are poorly understood but are similar to what occurrs during meiosis and mitosis when homolgous chromosomes align along the metaphase plane, the engineered construct finds the targeted gene and recombination takes place within the homolgous (meaning identical in this case) sequences (figure 3). The recombination may take place anywhere within the flanking DNA sequences and the exact location is determined by the cells and not the investigators.

Figure 3. Diagram of alignment of DNA just prior to homolgous recombination. Amazingly, the DNA construct finds its way into the nucleus and then aligns itself with the targeted locus. The mechanism that performs this alignment is poorly understood but it does work better in some species than others. One of the best species for performing homologous recombination is yeast. Mouse is a good mammalian system because homologous recombination does not work as eifficiently in human cells.
Once the cells have performed their part of the procedure, the end result is a new piece of DNA inserted into the chromosome. The rest of the genome is unaltered but the single targeted locus has been replaced with the engineered construct and some of its flanking DNA (figure 4). The original engineered construct has taken up the targeted gene of interest but since it cannot replicate in a nucleus, it is lost quickly in dividing cells while the modified chromosome replicates faithfully, including its new insert.
Cells that have undergone homologous recombination can be selected by addition of antibiotic to the growth medium (positive selection). Notice that the negative selection marker is not incorporated into the chromosome by homologous recombination.

Figure 4. Diagram of the final products of homologous recombination. The chromosome now contains a portion of the flanking DNA as well as all of the engineered construct which has taken the place of the original allele. The original allele has been recombined into the construct and thus is lost over time. From this point on, the cell will replicate the engineered construct as faithfully as any other portion of the chromosome.
If the targeting vector aligns in a non-homologous region of the genome, then recombination is random and the negative selection marker may become incorporated into the genome (figure 5).

Figure 5. Alignment of DNA just prior to non-homolgous recombination. In this case, the recombination will occur randomly. Notice that this time the tk marker will be included in the recombination event.
The final product of non-homologous recombination (figure 6) can survive positive (antibiotic) selection. However, there is a drug called gancyclovir that will kill any cell that contains the tk gene. So cells undergoing homologous recombination are grown in antibiotic to select for recombination and gancyclovir to kill any cells that successfully conducted non-homologous recombination.

Figure 6. End products of non-homolgous recombination. The positive and negative selection markers are incorporated into the chromosome so gancyclovir will kill cells with modified chromosomes as shown here.
A knockout mouse has had both alleles of a particular gene replaced with an inactive allele. This is usually accomplished by using homologous recombination to replace one allele followed by two or more generations of selective breeding until an breeing pair are isolated that have both alleles of the targeted gene inactivaated or knocked out. Knock out mice allow investigators determine the role of a particular gene by observing the phenotype of individuals that lack the gene completely.
Step 1. Isolate developing embryo at blastocyst stage. This embryo is from a strain of mice with gray fur.
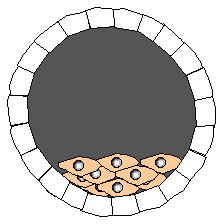
Step 2. Remove embryonic stem cells from gray-fur blastocyst. Grow stem cells in tissue culture.
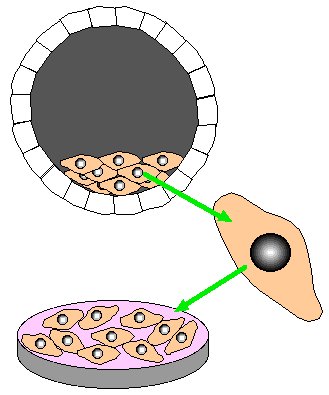
Step 3. Transfect stem cells with homologous recombination construct. Select for homologous recombination by growing stem cells in neomycin and gancyclvir.
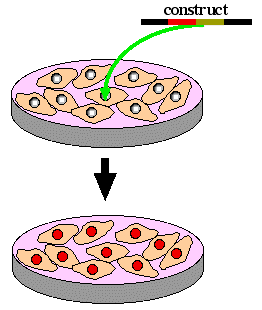
|
+ neomycin
|
+ gancyclovir
|
Step 4. Remove homologously recombined stem cells from petri dish and inject into a new blastocyst that would have only white fur.
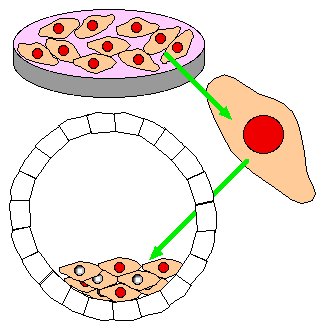
Step 5. Implant several chimeric blastocysts into pseudo-pregnant, white fur mouse.
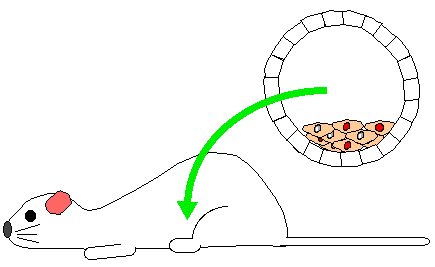
Step 6. Mother will give birth to a range of mice. Some will be normal white fur mice but others will be chimeric mice. Chimeric mice have many of their cells from the original white fur blastocyst but some of their cells will be derived from recombinant stem cells. Fur cells from recombinant stem cells produce gray patches which are easily detected.
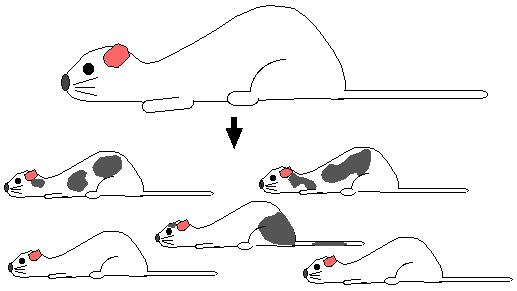
Step 7. Mate the chimeric mice with wild-type white fur mice. If the gonads of the chimeric mice were derived from recombinant stem cells, all the offspring will have gray fur. Every cell in gray mice are heterozygous for the homologous recombination.
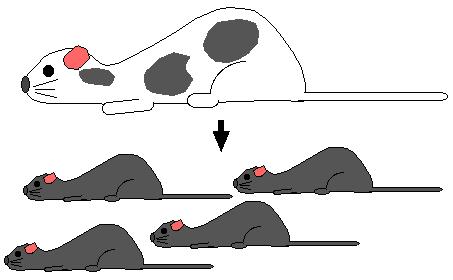
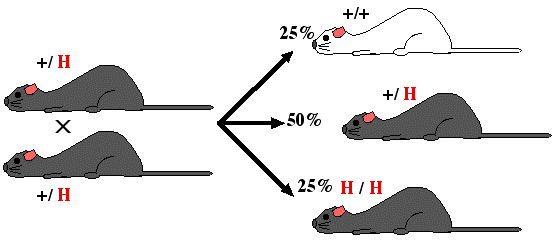
Step 8. Mate heterozygous gray mice (+/ H) and genotpye the gray offspring. Identify homozygous recombinants (H / H) and breed them to produce a strain of mice with both alleles knocked out. The pure breeding mouse strain is a "knockout mouse".
 knockout
mouse
knockout
mouse
© Copyright 2002 Department of Biology, Davidson College, Davidson,
NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu