This web page was produced as an assignment for an undergraduate course at Davidson College.
SST2 ~ My Favorite Yeast Gene
A little considered yeast (Sacchromyces cerevisiae) gene that
plays a primary role in GTPase activation.
The Background:
To begin to understand SST2, it's important to know about G proteins and the role they play in yeast reproduction. Dr.Janet Kurjan's home page provides a good overview of the role of G protein signalling and SST2's involvement. G proteins are signal transducers. In the case of SST2p (the SST2 protein), the yeast cell is responding to pheromones that initiate mating responses. SST2 functions to desensitize the signal response to these pheromones. Scientists are interested in the pattern of these responses because of the high degree of homology with mammalian RGS (regulators of G-proteins) genes associated with hemoglobin.Click here to see blast analysis results with mammalian homologies.
The best place to begin learning about my favorite yeast gene was its entry on the Sacchromyces Gene Database (SGD) maintained by Stanford University. Click here to view the entry. Summarily, the page reports that null mutants are viable and, in fact, do show increased sensitivity to mating factors (which is logical if the gene is associated with desentization to mating factors.) It's molecular function is GTPase activation, while its biological processes are described as signal transduction and adaptation to mating signal.
The Literature:
Dietzel and Kurjan (1987) initially closed the SST2 gene. They discovered the mutated copies of SST2 produced supersensitivity to both alpha and a-factor (yeast mating type proteins.) Their work placed SST2 in a signalling pathway that inhibits a pheromone response. Dohlman et al. (1996) discovered that cells require SST2p in order to resume growth after exposure to pheromone. Moreover, their work indicates a counterpart, Gpa1, that seems to physically interact with SST2. Together, they produce a negative feedback loop to pheromone exposure.
Apanovitch et al. (1998) examined the biochemical role of SST2. SST2 is the first gene in a new family of yeast genes, RGS (look above if this acronym is unfamiliar.) Its apparent role is desensitization to pheremone signalling. It appears that SST2 stimulates GTP hydroloysis, but does not regulate or modulate GTP hydrolysis. Because SST2 is associated with G protein signalling, Leberer et al. (1997) hypothesize that SST2 may be the alpha subunit on the GTPase-activating protein.
Location, location, location:
SST2 is located on Chromosome XII. The Stanford YGD can be accessed for an interactive look at the areas around the SST2 locus. The diagram below depicts the other loci surrounding SST2.
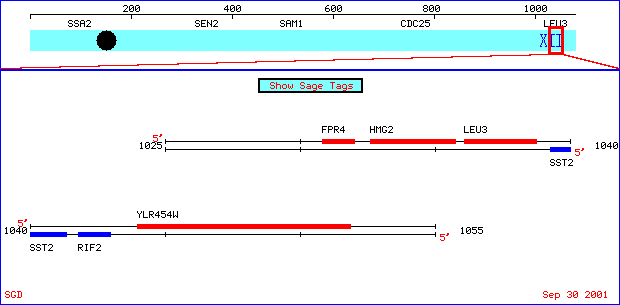
Structure:
SST2p has not yet been crystallized. Therefore, a PDB file is not available.
References:
Apanovitch DM, Slep KC, Dohlman HG. (1998) Sst2 is a GTPas-activating protein for Gpa1: purification and characterization of a cognate RGS-Galpha protein pair in yeast. Biochemistry. 37(14): 4815-22.
Dietzel C, Kurjan J. (1987) Pheromonal regulation and sequence of the Sacchromyces cerevisae SST2 gene: a model for desensitization to pheromone. Molecular Cell Biology. 7(12): 4169-77.
Dohlman HG, Song J, Ma D, Courchesne WE, Thorner J. (1996) Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical interaction with Gpa1 (the G-protein alpha subunit.) Molecular Cell Biology. 16(9): 5194-209.
Leberer E, Thomas DY, Whiteway M. (1997) Phereomone signalling and polarized morphogenesis in yeast. Current Opinions in Genetic Development. 7(1): 59-66.
http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=SST2. SGD entry for SSt2. Last accessed on October 1, 2001 11:00 a.m.
Email me at: amhartman@davidson.edu
Return to my home page
Return to the Davidson College Biology Department