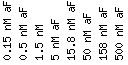|
This web page was produced as an assignment for an
undergraduate course at Davidson College. |
My Favorite
Yeast Gene: Microarray Data Analysis
This page builds on the work completed in my second web page, My Favorite Yeast Gene. Using my annotated (RAS1) and unannotated (YMR086W) genes, I use microarray data in this page to try and assign a function to both of the genes. Assigning function to RAS1 is slightly easier as experimentation has already been carried out to decipher that it has a role in determining the resources that are used in metabolic capacity and response to stress, and as a life span regulator (Shama 1998). YMR086W on the other hand, has no direct experimentation related with its function, and thus is a gene ready to be exposed.
Using the Expression Connection database from Stanford University, microarray data can be analyzed by looking at the grouping of genes under similar experimental conditions. These experiments were carried out by multiple groups, and are listed below followed by the group responsible for the work. To discover more about each particular experiment, click on the link below:
·
Expression
at different alpha-factor concentrations (Rosetta Inpharmatics)
·
Expression in response
to alpha-factor (Rosetta Inpharmatics)
·
Expression
in response to DNA-damaging agents (Stanford University)
·
Expression during the cell
cycle (Stanford University)
·
Expression during the diauxic
shift (Stanford University)
·
Evolution of expression during
glucose limitation (Stanford University)
·
Expression during
sporulation (Stanford University)
·
Expression in response to
environmental changes (Stanford University)
·
Expression
in response to histone depletion (The Whitehead Institute)
For Both RAS1 and YMR086W, not all of the above experiments produced useful results. Those that did (and in some cases those that didn’t) are shown below followed by an analysis of what the microarray data signifies about the genes function.
****************************************************************
All data use the following scale:
Scale : (fold repression/induction)
![]()
Click on a color strip to see data for that gene.
****************************************************************
Annotated: RAS1
Expression
at different alpha-factor concentrations for RAS1/YOR101W
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
RAS1 |
|
|
RAS protein signal
transduction* |
|
RAS small monomeric GTPase |
|
plasma membrane |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
RHK1 |
|
|
protein amino acid
glycosylation |
|
not yet annotated |
|
not yet annotated |
||
|
|
ARO9 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
PAD1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
MAM33 |
|
|
respiration |
|
not yet annotated |
|
not yet annotated |
||
|
|
TFC7 |
|
|
transcription initiation,
from Pol III promoter |
|
RNA polymerase III
transcription factor |
|
transcription factor TFIIIC |
||
|
|
FTR1 |
|
|
transport |
|
not yet annotated |
|
not yet annotated |
||
|
|
OLE1 |
|
|
mitochondrion inheritance* |
|
stearoyl-CoA desaturase |
|
endoplasmic reticulum
membrane |
||
|
|
PHO5 |
|
|
phosphate metabolism |
|
acid phosphatase |
|
periplasmic space (sensu
Fungi)* |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
ARO10 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
AST1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
PRP28 |
|
|
lariat formation, 5'-splice
site cleavage |
|
ATP dependent RNA helicase* |
|
spliceosome |
||
|
|
ELM1 |
|
|
protein amino acid
phosphorylation* |
|
protein kinase |
|
cytokinetic ring (sensu
Saccharomyces) |
||
|
|
RRP45 |
|
|
35S primary transcript
processing* |
|
3'-5' exoribonuclease |
|
nuclear exosome (RNase
complex)* |
||
|
|
RNH1 |
|
|
cell wall organization and
biogenesis* |
|
ribonuclease H |
|
cell |
||
|
|
SIT1 |
|
|
transport |
|
not yet annotated |
|
not yet annotated |
* : indicates that more than one annotation exists for the gene.
See the Summary of the Gene Ontology annotations for this gene group
Table taken from SGD
Expression Connection data base at the the following link: http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl?query=RAS1&noSearch=1
- In
this experiment, transcript profiling was used to monitor signal
transduction during yeast pheromone response (Roberts 2000). RAS1 was neither repressed nor
induced. The genes that RAS1
clustered with (those whose biological process is known) have varying
function, but many are involved in regulating cell morphology, growth, and
differentiation, thereby supporting work completed by Miller, et al in
1997.
Expression in response to alpha-factor for RAS1/YOR101W
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
RAS1 |
|
|
RAS protein signal transduction* |
|
RAS small monomeric GTPase |
|
plasma membrane |
||
|
|
RPC25 |
|
|
transcription, from Pol III promoter |
|
DNA-directed RNA polymerase III |
|
DNA-directed RNA polymerase III |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SNF6 |
|
|
chromatin modeling |
|
non-specific RNA polymerase II transcription factor |
|
nucleosome remodeling complex |
||
|
|
UTR2 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
TAF25 |
|
|
transcription initiation, from Pol II promoter* |
|
general RNA polymerase II transcription factor |
|
TFIID complex* |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
ALG11 |
|
|
protein amino acid glycosylation |
|
alpha-1,2-mannosyltransferase |
|
endoplasmic reticulum* |
||
|
|
HUT1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
NUP188 |
|
|
mRNA-nucleus export* |
|
structural protein |
|
nuclear pore |
||
|
|
RPL21B |
|
|
protein biosynthesis |
|
structural protein of ribosome |
|
cytosolic large ribosomal (60S) subunit |
||
|
|
TIF5 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
RPC19 |
|
|
transcription, from Pol I promoter* |
|
DNA-directed RNA polymerase III* |
|
DNA-directed RNA polymerase III* |
||
|
|
CDC6 |
|
|
pre-replicative complex formation and maintenance |
|
adenosinetriphosphatase* |
|
pre-replicative complex |
||
|
|
PNT1 |
|
|
inner mitochondrial membrane organization and biogenesis |
|
molecular_function unknown |
|
mitochondrial inner membrane |
||
|
|
RSC4 |
|
|
chromatin modeling |
|
molecular_function unknown |
|
nucleosome remodeling complex |
||
|
|
ORC6 |
|
|
DNA replication initiation* |
|
DNA replication origin binding |
|
origin recognition complex |
||
|
|
RPL30 |
|
|
protein biosynthesis* |
|
structural protein of ribosome |
|
cytosolic large ribosomal (60S) subunit |
||
|
|
RPS31 |
|
|
protein biosynthesis* |
|
structural protein of ribosome |
|
cytosolic small ribosomal (40S) subunit |
Table taken
from SGD Expression Connection data base at the the following link: http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl?query=RAS1&noSearch=1
·
In the above microarray data, the same alpha factor was tested, but
this time over length of time as opposed to amount of pheremone. Here, RAS1 was repressed after about 100
minutes, and clustered with genes which often influenced protein
biosynthesis. Following Miller’s
hypothesis of the regulatory function of RAS1 in the distributin of resources,
it makes sense that protein synthesis genes are repressed at the same time as
RAS1 is repressed, as RAS1 would normally cause that gene to be activated to
meet cell needs.
Expression in response to DNA-damaging agents for RAS1/YOR101W
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
RAS1 |
|
|
RAS protein signal transduction* |
|
RAS small monomeric GTPase |
|
plasma membrane |
||
|
|
RKI1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
PRS4 |
|
|
histidine biosynthesis* |
|
ribose-phosphate pyrophosphokinase |
|
cytoplasm |
||
|
|
AAH1 |
|
|
adenine catabolism |
|
adenine deaminase |
|
intracellular |
||
|
|
RPA49 |
|
|
transcription, from Pol I promoter |
|
DNA-directed RNA polymerase I |
|
DNA-directed RNA polymerase I |
||
|
|
MES1 |
|
|
not yet annotated |
|
methionine--tRNA ligase |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
NIP7 |
|
|
ribosomal large subunit assembly and maintenance* |
|
protein binding |
|
cytosolic large ribosomal (60S) subunit* |
||
|
|
ROK1 |
|
|
mRNA splicing* |
|
ATP dependent RNA helicase |
|
nucleolus |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SUP45 |
|
|
protein synthesis termination |
|
translation release factor, codon specific |
|
cytosol |
||
|
|
RPC40 |
|
|
transcription, from Pol I promoter* |
|
DNA-directed RNA polymerase III* |
|
DNA-directed RNA polymerase III* |
||
|
|
GCD7 |
|
|
protein synthesis initiation |
|
translation initiation factor |
|
ribosome |
||
|
|
SUI2 |
|
|
protein synthesis initiation |
|
translation initiation factor |
|
ribosome |
||
|
|
ELP2 |
|
|
transcription |
|
not yet annotated |
|
not yet annotated |
||
|
|
BRX1 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
HMT1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
* : indicates that more than one annotation exists for the gene.
Table taken from SGD Expression Connection data base at the the following link: http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl?query=RAS1&noSearch=1
- Here, when the genome was exposed to DNA damaging agents, RAS1 clustered with genes that regulate the initiation and termination of protein synthesis. Again, this supports the hypothesis that RAS1 functions in distributing resources around the cell. When RAS1 is repressed, so are these genes. It seems likely that RAS1 has a role in promoting these genes; with RAS1 repressed, they too are repressed.
 (Data Strips—Black, Green, Red—Are NOT Active)
(Data Strips—Black, Green, Red—Are NOT Active)
|
Name |
Score |
Peak |
|
|
0.292 |
|
|
|
|
Reference Genes |
|
||
|
10.9 |
G1 |
|
|
|
10.68 |
S |
|
|
|
3.08 |
G2 |
|
|
|
6.726 |
M |
|
|
|
11.8 |
M/G1 |
|
|
|
|
|
|
|
|
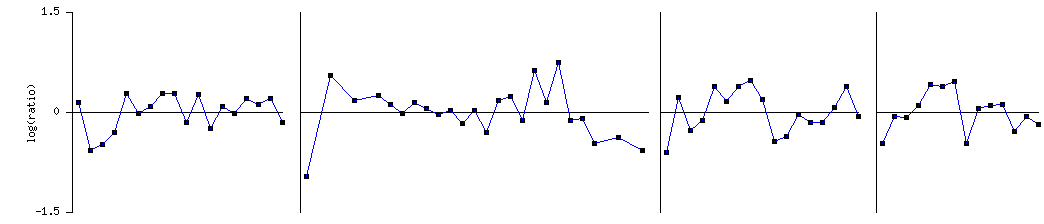
Plot of RAS1 (YOR101W) |
|
||
Top of Form