This web page was produced
as an assignment for an undergraduate course at Davidson College.
MY FAVORITE YEAST GENES
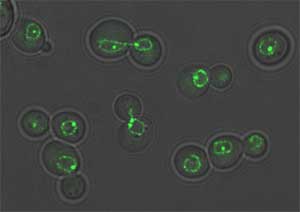
Figure 1. These budding yeast cells have been tagged
using GFP (Green Fluorescent Protein) and are visualized using fluorescence
microscopy (http://www.unige.ch/sciences/biologie/bimol/htmls/gasser-f.html).
Permission requested from Susan Gasser.
Annotated
Gene: FAR1
Unannotated Gene:YBR293W
FAR1 Relative Sites
References
Annotated Gene:
FAR1
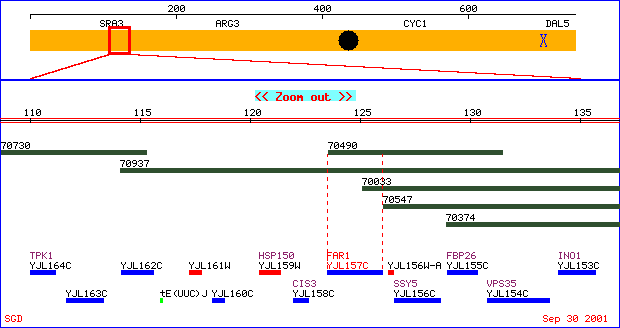
Figure 2. A physical map of FAR1 located on chromosome X.
The yellow depicts chromosome X. The blue designates a Crick strand
ORF and red a Watson strand ORF. ATCC clones are represented in green.
Image taken from http://genome-www4.stanford.edu/cgi-bin/SGD/PHYSMAP/PHYSmap?seq=FAR1
Permission requested from SGD.
FAR1
is located in the genome of Saccharomyces cerevisiae commonly known
as baker's or budding yeast (SGD, 2001; http://genome-www.stanford.edu/Saccharomyces/).
This gene is situated on chromosome X and is called such because it is
a "Factor ARrest" gene (YPD, 2001; http://www.proteome.com/databases/YPD/reports/FAR1.html).
FAR1 is a cyclin-dependent protein kinase inhibitor involved in cell-cycle
arrest (SGD, 2001; http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=FAR1).
This arrest in the G1 phase is important because it allows for cell-cycle
synchronization between two haploid cells with opposite mating types.
The cell-cycle arrest begins with the secretion of peptide pheromones a
and
alpha
by haploid yeast cells. This secretion induces conjugation between
cells of opposite mating types along with morphological changes within
the cells when the pheromones bind to their receptors. This binding
initiates a signal transduction pathway since the receptor is coupled with
a heterotrimeric guanosine triphosphate-binding protein (G protein) in
both haploid yeast cells. A mitogen-activated protein (MAP) kinase
cascade is stimulated and Cdc28 protein kinase is inactivated (Leeuw et
al. 1210). The cell cycle is halted since Cdc28p is essential
for the G1 phase to end and the S phase to begin. Far1p inactivates
the Cdc28 protein kinase of the Cdc28p-Clnp complex. The transcription
of FAR1 is actually induced by the secreted mating pheromone (Peter and
Herskowitz 1230). FAR1 may also play a role in the polarized orientation
of growth during mating (Butty et al. 1511).
A FAR1 mutant has a viable phenotype. Two substitutions in FAR1 lead
to the far1-D1 mutation--glycine 646 and proline 671 are replaced by aspartic
acid and leucine, respectively (Figure 3). This null mutant can still
induce the signal transduction pathway as in the wild-type, but is unable
to halt the cell-cycle at the G1 phase when exposed to mating pheromone.
Far1-D1 mutant exposure to mating pheromone can also lead to unusual cell
morphology. However when these mutants are not affected by mating
pheromone, they have normal cell growth, budding, size, and pheromone production.
A second FAR1 mutant (far1-22p) results from the replacement of serine
87 with proline (Figure 3). This mutant can induce a G1 arrest in
the absence of a mating pheromone by altering the phosphorylation by the
Cdc28p-Cln2p kinase complex. There are six other null mutants including
FAR1-22,nls1; FAR1-22deltaNES, far1-s, Far1-60F3p, farc1-c, and far1-H7
at the FAR1 locus (YPD, 2001; http://www.proteome.com/databases/YPD/reports/FAR1.html).
1 MKTPTRVSFE KKIHTPPSGD
RDAERSPPKK FLRGLSGKVF
41 RKTPEFKKQQ MPTFGYIEES QFTPNLGLMM SKRGNIPKPL
81 NLSKPISPPP
SLKKTAGSVA SGFSKTGQLS ALQSPVNITS
121 SNKYNIKATN LTTSLLRESI SDSTTMCDTL SDINLTVMDE
161 DYRIDGDSYY EEDSPTFMIS LERNIKKCNS QFSPKRYIGE
201 KCLICEESIS STFTGEKVVE STCSHTSHYN CYLMLFETLY
241 FQGKFPECKI CGEVSKPKDK DIVPEMVSKL LTGAGAHDDG
281 PSSNMQQQWI DLKTARSFTG EFPQFTPQEQ LIRTADISCD
321 GFRTPRLSNS NQFEAVSYLD SPFLNSPFVN KMATTDPFDL
361 SDDEKLDCDD EIDESAAEVW FSKTGGEHVM VSVKFQEMRT
401 SDDLGVLQDV NHVDHEELEE REKEWKKKID QYIETNVDKD
441 SEFGSLILFD KLMYSDDGEQ WVDNNLVILF SKFLVLFDFE
481 EMKILGKIPR DQFYQVIKFN EDVLLCSLKS TNIPEIYLRF
521 NENCEKWLLP KWKYCLENSS LETLPLSEIV STVKELSHVN
561 IIGALGAPPD VISAQSHDSR LPWKRLHSDT PLKLIVCLNL
601 SHADGELYRK RVLKSVHQIL DGLNTDDLLG IVVVGRDGSG
641 VVGPFGTFIG
MINKNWDGWT TFLDNLEVVN PNVFRDEKQQ
681 YKVTLQTCER LASTSAYVDT DDHIATGYAK QILVLNGSDV
721 VDIEHDQKLK KAFDQLSYHW RYEISQRRMT PLNASIKQFL
761 EELHTKRYLD VTLRLPQATF EQVYLGDMAA GEQKTRLIM
801 EHPHSSLIEI EYFDLVKQQR IHQTLEVPNL
Figure 3. Wild-type amino acid sequence of Far1p.
The far1-D1 mutation results from two substitutions, G646D and P671L.
Glycine 646 and proline 671 are both blue.
The far1p-22 mutation results from a substitution of serine 87 (red)
to proline. Sequence taken from http://www.proteome.com/databases/YPD/reports/FAR1.html.
Relative Sites
Yeast
Proteome Database Protein Report for FAR1- This site is a wealth
of knowledge--it describes FAR1's function, pathway, mutant phenotype,
genetic interactions, etc.
SGD-
This site provides a summary of FAR1.
Annotated
Yeast Gene: CDC28- A fellow student's web page describing CDC28,
a cyclin-dependent kinase. This page describes the cell cycle and
the role of cyclin-dependent kinases.
Unannotated Gene: YBR293W
The unannotated gene YBR293W is an ORF (open reading frame) since only
physical properties are known.
It is located on the long arm of chromosome 2 in the genome of Saccharomyces
cerevisiae.
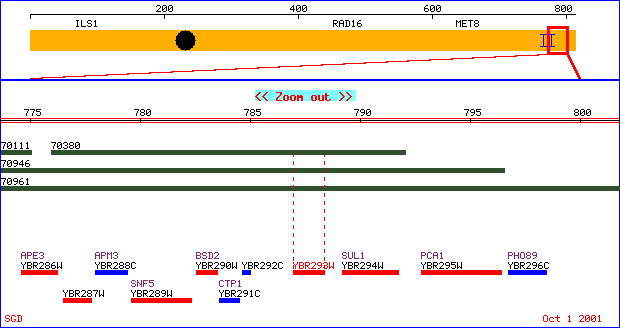
Figure 4. Physical map of chromosome 2 illustrating the unannotated
gene, YBR293W.
SGD-Nucleotide
Sequence
1 ATGAGTATTT CAAATTGGAT CACCACTGCG
TATTTAATTA CATCAACATC
51 TTTTCAACCT CTTTATGGGT CATTTTCTGA
TGCACTTGGT CGAAGAAACT
101 GCCTTTTCTT TGCTAATGGG GCTTTTACCA
TTGGATGTCT AGCCTGTGGT
151 TTCTCGAAAA ACATCTACAT GCTTAGTTTT
ATGAGAGCAT TGACAGGCAT
201 AGGAGGTGGT GGCTTGATCA CACTTTCTAC
AATCGTAAAT TCAGACGTTA
251 TTCCAAGTTC GAAAAGAGGA ATTTTTCAAG
CGTTTCAGAA TTTACTTTTG
301 GGATTTGGTG CCATATGTGG AGCGTCTTTC
GGTGGCACAA TAGCGTCGAG
351 CATTGGTTGG AGGTGGTGTT TTCTCATCCA
AGTACCCATA TCTGTGATTA
401 GTTCCATATT AATGAATTAT TATGTACCTA
ATCAGAAAGA ATATAATCGT
451 CAAAATTCTA GCATATTCCA AAATCCCGGA
AAAATACTCA GGGACATAGA
501 TGTTATGGGC TCAATTCTTA TTATAACTGG
TCTCACACTA CAGCTTCTTT
551 ACCTGAGCCT GGGGTGTTCT ACTTCTAAAT
TATCATGGAC CAGCCCTTCT
601 GTGCTACTGC TATTAGTTGG GAGTGTAATA
ATCCTCTTAC TGTTCATATT
651 GCACGAAAGG AAAACAAGTG CTAGAGCGAT
TATTCCTATG GAGCTGGTCA
701 ATTCCTCCTA CAGTGTCGTT GTACTTTCGA
TAAGTATACT TGTTGGTTTT
751 GCCAGCTACG CGTATCTTTT TACTTTACCA
TTATTCTTTC AGATTGTACT
801 TGGAGATTCC ACTGCAAAAG CAGGATTACG
TCTTACGATT CCTTCCCTAT
851 TTACTCCGGT AGGCAGTCTC ATAACAGGAT
TTTCCATGAG CAAGTACAAC
901 TGTCTAAGAT TATTACTCTA CATTGGTATT
TCTTTGATGT TTTTGGGTAA
951 CTTTTTATTC CTGTTTATTG AAAAAACTTC
TCCGAACTGG TTGATTGGTC
1001 TATTTTTGAT ACCTGCAAAT CTAGGACAAG GTATCACTTT
TCCTACGACC
1051 TTGTTTACTT TCATATTTAT GTTCTCTAAG AGTGACCAAG
CTACTGCGAC
1101 ATCAACTTTA TATTTATTCC GTAGTATTGG ATCTGTATGG
GGTGTTGCAA
1151 TTTCAGCTGG CGTCATTCAA TTATCTTTCG CAGGTTTATT
GCGTAGTAAT
1201 TTGAAAGGTC TACTGGATGA AAACAAGATA AAGAAACTTA
TTGTTCAGCT
1251 TAGTGCAAAC TCCTCATATA TTGGATCTTT ACATGGCGAA
GTTAAAAACA
1301 CAGTCATAAA GAGTTTTGAT GAGGCAACAA AGAGGGCTCA
TCTAATGTCT
1351 ACATTACTCT CTTCATTGGC CCTGATACTC TGCATCCTTA
AAGACAATCT
1401 GGCGAAACCT AAAACAAGAA GATAA
Nucleotide BLAST
species: Saccahromyces cervisiae
definition: chromosome II reading frame
locus: SCYBR293W
E=0.0, excellent match
nucleotide length: 4160 bp
accession number: Z36162 Y13134
gi: 536749
amino acid sequence: MSISNWITTAYLITSTSFQPLYGSFSDALGRRNCLFFANGAFTI
GCLACGFSKNIYMLSFMRALTGIGGGGLITLSTIVNSDVIPSSKRGIFQAFQNLLLGF
GAICGASFGGTIASSIGWRWCFLIQVPISVISSILMNYYVPNQKEYNRQNSSIFQNPG
KILRDIDVMGSILIITGLTLQLLYLSLGCSTSKLSWTSPSVLLLLVGSVIILLLFILH
ERKTSARAIIPMELVNSSYSVVVLSISILVGFASYAYLFTLPLFFQIVLGDSTAKAGL
RLTIPSLFTPVGSLITGFSMSKYNCLRLLLYIGISLMFLGNFLFLFIEKTSPNWLIGL
FLIPANLGQGITFPTTLFTFIFMFSKSDQATATSTLYLFRSIGSVWGVAISAGVIQLS
FAGLLRSNLKGLLDENKIKKLIVQLSANSSYIGSLHGEVKNTVIKSFDEATKRAHLMS
TLLSSLALILCILKDNLAKPKTRR
PREDATOR
(predicts secondary structure of a protein)
51.05% random coil
26.16% alpha-helix
22.78% extended strand
The reliability plot did not indicate that this
was a correct prediction since very little of it was above 0.8.
Kyte-Doolittle
Analysis (predicts whether protein is an integral membrane protein)
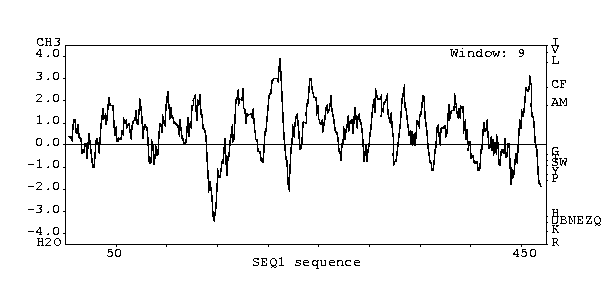
This ORF most likely codes for an integral membrane
protein since the hydropathy plot peaks above 2.0.
Conserved
Domain
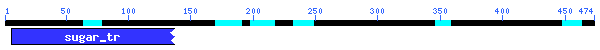
There was only 30.4% alignment with a sugar transporter with an E-value
of 1x10-4. This must not be a very highly conserved sequence.
YBR293W most likely codes for an integral membrane protein, but I could
not ascertain the function of this gene. The only prediction I can
make regarding the role of this gene in the life of yeast would be that
it is involved in budding since genes that regulate budding are not highly
conserved.
References
Butty AC, et al.1998. The Role of Far1p in linking the heterotrimeric
G protein to polarity establishment proteins during yeast mating. Science
282:1511-1516.
Kyte J, Doolittle
RF. 1982. J. Mol. Biol. 157:105-132.
Leeuw T, et al. 1995. Pheromone Response in Yeast: Association
of Bem1p with Proteins of the MAP Kinase Cascade and Actin. Science 270:1210-1213.
Peter M, Herskowitz I. 1994. Direct Inhibition of the Yeast Cyclin-Dependent
Kinase Cdc28-Cln by Far1. Science 265:1228-1231.
NCBI. BLASTn Results. http://www.ncbi.nlm.nih.gov/blast/Blast.cgi#536749
Accessed 2001 Sept 30.
NCBI. Conserved Domain Database. http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
Accessed 2001 Sept 30.
PREDATOR. PREDATOR result for UNK_2789840. http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa-predator.html
Accessed 2001
Sept 30.
SGD. FAR1/YJL157C. http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=FAR1
Accessed 2001 Sept 28.
YPD. YPD Protein Report for FAR1. http://www.proteome.com/databases/YPD/reports/FAR1.html
Accessed 2001 Sept 28.
Home
Genomics
Course
Davidson College
Email me!