*This page was produced as part of an undergraduate course at Davidson College*
My Favorite Yeast Genes
Annotated Yeast Gene- FUR4
What is FUR4?
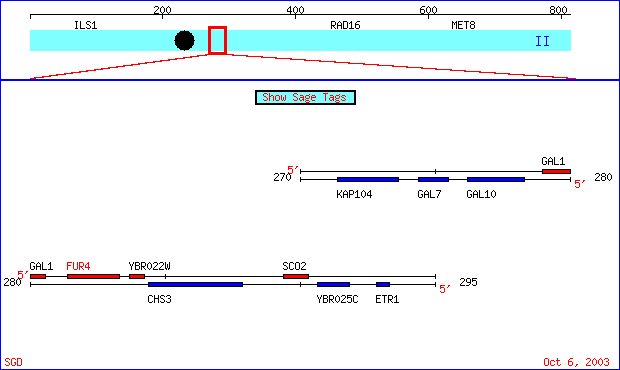
FUR4 is one of a family of genes that is found on chromosome 2 in Saccharomyces cerevisae. Seen below is an image of FUR4's relative location on chromosome 2 as well as several of the genes around it.

Figure 1- This is an image of a region of chromosome II from the 271402 to the 293303 bp of S. cerevisae taken from SGD. The FUR4 gene is highlighted in red. (SGD; 2003, <http://db.yeastgenome.org/cgi-bin/SGD/ORFMAP/ORFmap?sgdid=S0000225>)
The FUR4 gene amino acid sequence is as follows according to the NCBI Entrez-Protein database. (NCBI; 2003, <http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?cmd=Retrieve&db=protein&list_uids=536226&dopt=GenPept&term=&qty=1>):
1 mpdnlslhls
gsskrlnsrq lmessnetfa pnnvdlekey kssqsnitte vyeassfeek
61 vssekpqyss fwkkiyyeyv vvdksilgvs
ildsfmynqd lkpvekerrv wswynycyfw
121 laecfnintw qiaatglqlg lnwwqcwiti wigygfvgaf
vvlasrvgsa yhlsfpissr
181 asfgiffslw pvinrvvmai vwysvqayia atpvslmlks
ifgkdlqdki pdhfgspnat
241 tyefmcffif waaslpfllv pphkirhlft vkavlvpfas
fgfliwairr ahgrialgsl
301 tdvqphgsaf swaflrslmg cmanfstmvi napdfsrfsk
npnsalwsql vcipflfsit
361 cligilvtaa gyeiyginyw spldvlekfl qttynkgtra
gvflisfvfa vaqlgtnisa
421 nslscgtdms aifpkfinik rgslfcaama lcicpwnlma
tsskftmals ayaiflssia
481 gvvcsdyfvv rrgyiklthi yshqkgsfym ygnrfginwr
alaaylcgva pclpgfiaev
541 gapaikvsdg amklyylsyw vgyglsfssy talcyffpvp
gcpvnniikd kgwfqrwanv
601 ddfeeewkdt ierddlvddn isvyehehek tfi
Figure 2- The amino acid sequence of the FUR4 gene.
FUR4 encodes for a protein known as uracil permease. A search of the PDB (PDB [protein data bank]; 2003, <www.pdb.org>) yielded no structures for uracil permease. As the name suggests, it is associated with the pyrimidine, uracil.
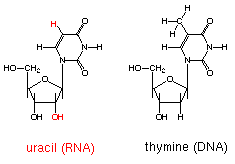
Figure 3- A simple image of uracil in one of its roles as an RNA nucleotide.
*Image permission pending from the biology department at MIT*
Uracil was first isolated in the early 20th century from herring sperm cells and it has many roles. It has many different derivatives, the most important of which is probably UTP (uridine triphosphate). This uracil derivative is used in carbohydrate metabolism by acting as a coenzyme in the biosynthesis of critical sugars across many different organisms. It can also be converted into ATP, that all important cellular energy source, by donating one of its phosphate groups. Perhaps most importantly, uracil has a crucial role in genetics as it is a component of RNA in complimentary pairing with adenine. (The preceding information on Uracil was taken from The Columbia Encyclopedia at Bartleby; 2003, <http://www.bartleby.com/65/ur/uracil.html>)
Let us shift our attention back to the focus of this webpage. Why is it that uracil and uracil permease, the product of FUR4, are associated with one another?
Biological Process-
The role of FUR4 is to produce uracil permease, as mentioned before, which is a specific co-transporter used in yeast cells to move uracil into and out of cells across the normally nearly impervious cell membrane. It is a membrane protein spanning the membrane 10 times that is delivered through a typical secretory pathway (Urban et al., 1999). Uracil permease is similar to many other transporter proteins such as lactose permease or alanine permease in both function and structure. The most important part of the protein is the hydrophobic core, which contains three charged residues in its transmembrane domain. These residues include two glutamic residues at positions 243 and 539 and a lyseine residue at position 272, their location within the core suggest they are involved in transporter structure and function within a hydrophobic environment (Urban et al., 1999).
Molecular Function-
Uracil permease is a symporter enzyme nestled within the plasma membrane. It functions by generating an electrochemical gradient across the cell membrane that is selectively permeable to uracil and by binding its substrate then releasing it within the membrane. The gradiant in this scenario is generated by an active co-transport of protons within the protein at the same time that the uracil is transported (Urban et al., 1999). The charged residues within the permeases hydrophobic core definitely have a lot to do with this functioning as one of the most documented mutations suggets. In a study done by Urban et al. in 1995 showed that replacement of Lys-272 by Glu (K272E) greatly impairs the function of uracil permease through alteration of the binding site for uracil in the protein. This mutation did not have an effect on the folding pattern of the protein and resulted in a viable phenotype with drastically reduced amounts of uracil uptake (Pinson et al., 1995).
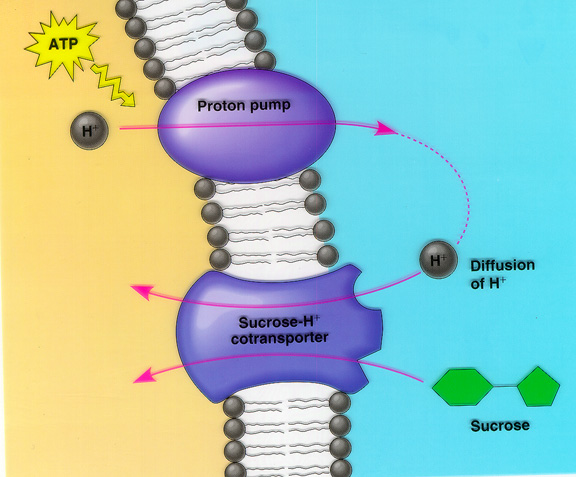
Figure 4- A typical diagram of co-transport within a cell as it occurs with the Sucrose-H+ cotransporter. Something of similar mechanism happens with uracil permease.
*Image permission pending from Miami U*
Cellular Component-
As mentioned before, the uracil permease protein produced by FUR4 is an integral membrane protein. Its location in the membrane is crucial and paramount to its function. Like other nucleotide transporters, it is specifically tailored chemically to transport uracil across the plasma membrane. I examined a Kyte-Doolittle Hydropathy plot to verify its properties as a membrane protein.

Figure 5- This is a hydropathy plot indicating that uracil permease is in fact integral to the membrane with 10 portions spanning the membrane. Notice the 10 portions of the line with a score greater than 1.8. (Kyte-Doolittle Hydropathy Plot; 2003, <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm>)
Un-Annotated Gene- YBLO10C
My favorite un-annotated gene about which only the amino acid sequence is truly known is YBLO10C, it is located on chromosome 2 along with FUR4. The region it is in can be viewed in the image below:
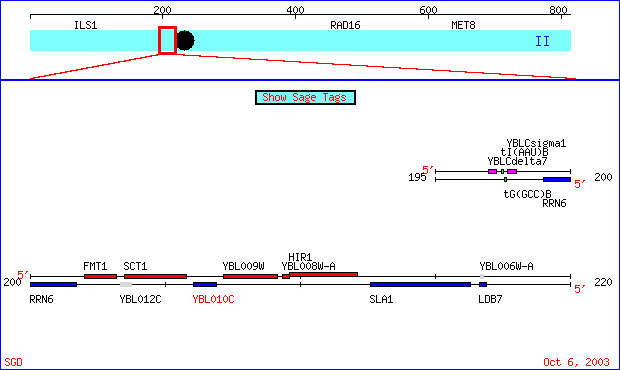
Figure 6- This is a region of chromosome 2 from the 196068 to the 216910 bp taken from SGD (SGD; 2003, <http://db.yeastgenome.org/cgi-bin/SGD/ORFMAP/ORFmap?sgdid=S0000106>). The gene is highlighted in red.
The amino acid sequence of this un-annotated gene according to NCBI Entrez-protein is (NCBI; 2003, <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=535996&dopt=GenPept>):
1 msdrdqiepv
tnaldaesds sddfgnfsda svendlynqn stlttssesv vdnclnkilp
61 kgefdleeet ikndcfklsk liederphvi
yeqlvqldpv lqpfiwnksh irrnllhilr
121 lsdnngsegv gtkreeepln delfkricda vekneqtatg
lflrdnfkid ytppmtlksl
181 qkeeereqeq hipqllmadf tsmdeeslrq yhdtlcqsid
flvsksrslk kqqrdllkdk
241 ttfenvvtnl tghtqrlqrd eialynkkrn kkkrfswvgy
Figure 7- The amino acid sequence of the YBL010C gene.
The first thing that I did was to do a Blastp search with the amino acid sequence to see which proteins on record that this amino acid sequence resembles. The closest match that was not the protein itself was the following (NCBI; 2003, <http://www.ncbi.nlm.nih.gov/blast/Blast.cgi>)
>gi|22977675|ref|ZP_00023463.1| COG0112: Glycine/serine hydroxymethyltransferase [Ralstonia
metallidurans]
Length = 566
Score = 36.6 bits (83), Expect = 0.52
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query: 155 EQTATGLFLRDNFKIDYTPPMTLKSLQKEEEREQEQHIPQLLMADFTS 202
+QT+ +F R F I+ P ++QKE +R QE HI + ++TS
Sbjct: 146 KQTSHAMFERSRFTIEQIDPEVFAAIQKENQR-QEDHIELIASENYTS 192
Figure 8- The nearest known protein in the BLASTp search.
This protein was not, to me, convincingly similar; however, it is intriguing to keep in mind that it was most common to a bacterial glycine/serine hydroxymethyltransferase. Next, I ran the amino acid sequence through the Kyte-Doolittle Hydropathy plot tool and obtained the following result:
Figure 9- The Kyte-Doolittle hydropathy plot results for YBL010C. This suggests it is not a transmembrane protein. (Kyte-Doolittle Hydropathy Plot; 2003, <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm>)
There were no spikes in hydropathy score over 1.8 and thus this protein is not functioning within the membrane. Next, I decided to test the protein that was found earlier in the BLASTp by running a Kyte-Doolittle hydropathy plot. I found that unlike the unknown protein, this protein did have a few spikes with hydropathy score higher than 1.8 which suggests a drastically different function and most likely invalidates any hopes of the two proteins having similar function.
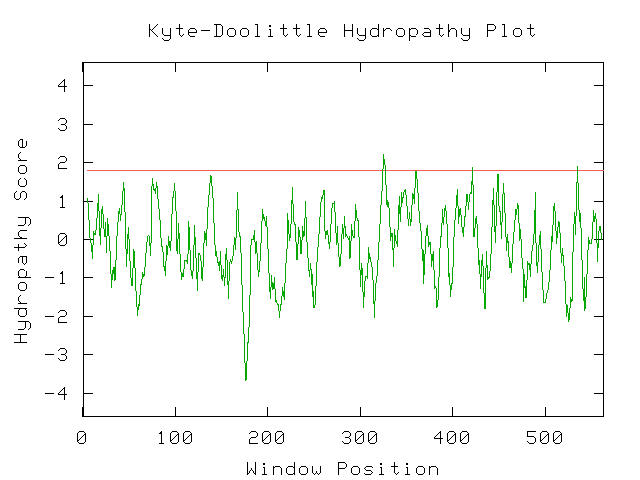
Figure 10- The Kyte-Doolittle Hydropathy results for COG0112. As can be seen, a few points with a score of higher than 1.8 are present suggesting that this protein is associated with a membrane function. (Kyte-Doolittle Hydropathy Plot; 2003, <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm>)
As there were no proteins in the BLASTp to compare this unknown protein to, it is difficult for me to make any conclusion about its cellular role. I also feel that it most likely has something to do with activity in or around the nucleus due to a combination of its low hydropathy score and the DNA-binding motif in the protein that can be seen by using the Swiss-Prot database.
1
11 21
31 41
51
1 MSDRDQIEPV TNALDAESDS SDDFGNFSDA SVENDLYNQN STLTTSSESV
VDNCLNKILP 60
61 KGEFDLEEET IKNDCFKLSK LIEDERPHVI YEQLVQLDPV LQPFIWNKSH
IRRNLLHILR 120
121 LSDNNGSEGV GTKREEEPLN DELFKRICDA VEKNEQTATG LFLRDNFKID YTPPMTLKSL
180
181 QKEEEREQEQ HIPQLLMADF TSMDEESLRQ YHDTLCQSID FLVSKSRSLK
KQQRDLLKDK 240
241 TTFENVVTNL TGHTQRLQRD EIALYNKKRN KKKRFSWVGY
Figure 11- Further analysis of the amino acid sequence of YBL010C taken from the Swiss-Prot database. The DNA-binding motif is highlighted in red (Swiss-Prot; 2003, <http://us.expasy.org/cgi-bin/sprot-ft-details.pl?P32788@DNA_BIND@225@240>)
References-
Bartleby; 2003, <http://www.bartleby.com/65/ur/uracil.html
Kyte-Doolittle Hydropathy Plot; 2003, <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm>
NCBI; 2003, <http://www.ncbi.nlm.nih.gov/blast/Blast.cgi>
Pinson, B et al. Only one of the charged amino acids located in membrane-spanning regions is important for the function of the Saccharomyces cerevisiae uracil permease. Biochem J. 1999 Apr 1;339 ( Pt 1):37-42.
PDB [protein data bank]; 2003, <www.pdb.org>
SGD; 2003, <http://db.yeastgenome.org/cgi-bin/SGD/
Urban, Grimal D et al. Replacement of Lys by Glu in a
transmembrane segment strongly impairs the function of the uracil permease from
Saccharomyces cerevisiae. Biochem J. 1995 Jun 15;308 ( Pt 3):847-51.
Return to Matt's Genomics Homepage
Return to the Genomics Homepage
Return to the Davidson Homepage
Email the Webmaster- matalbert@davidson.edu
.