My Favorite Yeast Expression
AFR1 & YDR089W
This web page was produced as an assignment for an undergraduate course
at Davidson College.
Assignment
Using microarray data to examine the expression profiles of the two genes from "My Favorite Yeast Genes," I hope to either support my hypohesis that YDR089W is involved in the extension of the membrane during shmoo formation and alpha-factor reception or reject and reformulate my hypothesis. In order to do so I will compare the expression profiles of AFR1 (known function) to YDR089W (unknown function), examining under which each is induced or repressed.
AFR1

(SGD, 2006; http://db.yeastgenome.org/cgi-bin/expression/expressionConnection.pl?locus=YDR085C&type=summary)
YDR089W

(SGD, 2006; http://db.yeastgenome.org/cgi-bin/expression/expressionConnection.pl?locus=YDR089W&type=summary)
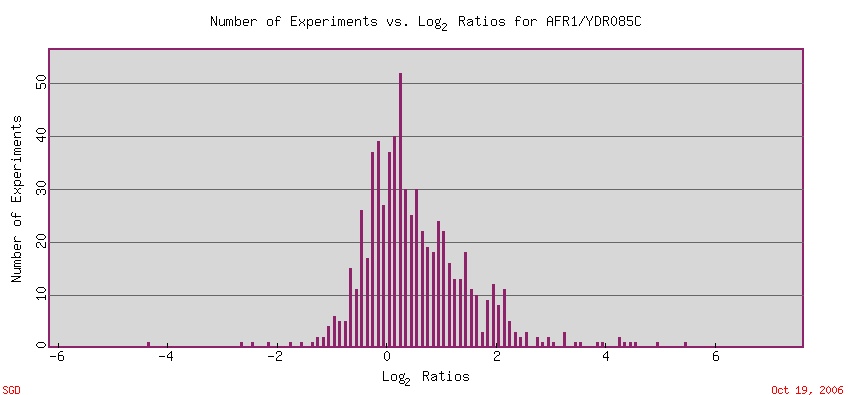
Based on the two figures above, at first glance AFR1 and YDR089W boh appear to have relatively similar graphs of number of experiments plotted against the respective Log2 ratios. Both tend to exhibit the standard bell-shapred curve, with the majority of experiments for each gene occuring near 0. With further examination, however, one can notice several interesting factors. The peak for number of experiments for the two graphs are different with YDR089W's being about 20 higher. More experiments produced results for YDR089W having a Log2 ratio near 0 than for AFR1. More experiments have produced results for AFR1 having a Log2 ratio further away from 0 than for YDR089W. Also, AFR1's curve appears to have more data in the positive Log2 ratios, whereas the YDR089W curve appears to have more in the negative Log2 ratios.
AFR1 and YDR089W Compared
The following scale is valid for all of the expression profile figures displayed on this page. As one can see, green dye denotes gene repression whereas red dye denotes gene induction. The intensity of the fluorescent dyes is directly proportional to the intensity of the repression or induction. Areas of the figures where the ratio of repression to induction is 1:1 appear black; this implies a lack of change in the gene's transcription between the control and experimental conditions (Campbell et al., 2006).

(SGD, 2006; http://db.yeastgenome.org/cgi-bin/expression/expressionConnection.pl?locus=YDR085C&type=summary)
In order to better understand the possible function of YDR089W, I decided it best to analyze microarray data in which AFR1 and YDR089W exhibited opposite expression patterns. The primary reason for this action was a general lack of similar expression patterns between the two genes, at least as far as the Saccharomyces Genome Database's Expression Connection presented. This fact alone hints toward a more likely rejection of my previous hypothesis.
Microarray and Expression Data
In 2004, research published in an article by Cullen et al. demonstrated the effects that glycosylation defects had on yeast's gene expression levels (SGD, 2006). Although not at all the focus of the paper, the microarray and expression profile data included the genes AFR1 and YDR089W. The following image shows the expression of both genes under their experimental conditions. As one should notice, there is a distinct opposite expression for the two genes under the condition of -/+ mannose.

Although these results do show that YDR089W is expressed differently than AFR1 in the conditions for Cullen et al.'s experiment, it might be more beneficial to compare the expression profiles for experiments that may deal more specifically with AFR1's function, cellular component, pathway, etc. On such study published in an article by Roberts et al. looked at yeast genes' expression levels as they responded to alpha-factor over time (SGD, 2006).

(SGD, 2006; http://db.yeastgenome.org/cgi-bin/expression/expressionConnection.pl?dataset=alpha_time&id=1125&type=enlarge&expts=7)
As one would hopefully assume, AFR1's expression was induced very strongly over time, seeing as AFR1 encodes the cytoskeletal protein which serves as the regulator for the alpha-factor receptor. Interestingly enough, not only was YDR089W not induced as well, but it was very strongly repressed. Its repression seems to have peaked in the 45 to 60 minute range of the experiment, whereas AFR1's induction remained at a ratio of greater that 2.8:1 beyond the 15 minute mark. This is considerable evidence to suggest that the two genes do not share similar functions within yeast. Further evidence can be found by examining the results, published in the same article, that examined the yeast's gene expression levels as they responded to varying concentrations of alpha-factor (SGD, 2006).

Just as in the previous set of microarray data, one sees AFR1 induced and YDR089W repressed. Given the experimental conditions of the study, the two genes appear to be expressed inversely. So, perhaps YDR089W is not involved in the membrane extension during shmooing and alpha-factor reception, but it obviously shuts off during this cellular process. Thus my hypothesis must be rejected, although the two genes are apparently related on some level. Considering that the genes are found at nearby locations it is interesting that they would be expressed inversely during a condition directly involving one of them (AFR1). Perhaps YDR089W is responsible for encoding a protein that must/should be "turned-off" in order for alpha-factor reception and shmooing to occur effectively. To examine things from a slightly different angle, I decided to look for a study with results where AFR1 was repressed under a condition or conditions for which YDR089W was induced. This was rather difficult; the only good data which I found was the following, from a study by Chu et al. examining gene expression during yeast sporulation:
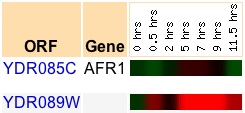
(SGD, 2006; http://db.yeastgenome.org/cgi-bin/expression/expressionConnection.pl?dataset=sporulation&id=1125&type=enlarge&expts=7)
Looking at this data, one sees that exist the same inverse relationship between the expression of the two genes. However, it is interesting to note the inverse at 0.5 hours with the repression of AFR1 and the induction of YDR089W. Also, YDR089W is several times more induced than is AFR1 from 2 hours to 9 hours. The two genes pick up the inverse expression again at 11.5 hours. I found this data interesting, given that sporulation and alpha-factor reception would both be involved in the overall category of reproduction.
Given this information, I would propose a new hypothesis that YDR089W, although not involved in membrane extension during shmooing and alpha-factor recption, encodes a membrane protein involved in the sporulation process.
References
Links
Genomics
Page
My
Home Page