AFR1 & YDR089W
This web page was produced as an assignment for an undergraduate course at Davidson College.
http://db.yeastgenome.org/cgi-bin/gbrowse/scgenome/
Annotated Gene: AFR1
- Description - The AFR1 gene in Saccharomyces cerevisiae encodes the cytoskeletal protein which serves as the regulator for the alpha-factor receptor. In yeast, the alpha-factor receptor plays an integral role in the mating process by receiving and binding the alpha-factor pheromone in the first stages of a G-protein-coupled signal transduction pathway. Ultimately, this pathway allows for the effective mating and shmooing (localized elongation of the cell wall for mating projection formation). The AFR1 gene encodes the principle protein which negatively regulates this pathway and thus controls, to at least a certain extent, the mating process of the yeast.
Gene Ontology:
Molecular Function - receptor signaling protein activity
Biological Process - cellular morphogenesis during mating, pheromone-dependent signal transduction during mating, and regulation of G-protein-coupled STP.
Cellular Location - mating projection base
Location:
Chromosome IV, base pairs 622109 to 624718
- Amino Acid Sequence Information
(from NCBI)
ORIGIN
001 megsylsaqe nqpiperlip rsnstsnlfa lsstfsklnv rndadynysn pnkkrhiysg
061 eidcrsvtaa rkfpvrscsm taaqqrkrta lftvrernsy hegfnndqdy vsqyqkpqyt
121 fgvykeltpy qlqrskmkrs fqfpngeiyk pkldgkcths lkkpelnsrd sslfkfsekk
181 grnlskdfvg phngtsvihi ppndtgygvn slelntsvps tikssvssts pisavntlts
241 lpesqtdddd gyenktvtis ycfentvnek hgshiekldl stkektkptt nsglfdrkkk
301 tilgtekyrc iksqsklklg svlkklwrts gnsntkhgkk dtkrrripid dmvthsdgns
361 eaendielmd anldgiefdd detlmdtdsi fddllskend kydlrrrqle irqklhetsh
421 nddgkvsfrd tekhnvnegl idktiieefs klgeyiidtr nqppprsskr pslddnesar
481 yfynistdlr qslsgpislp mhvgndmvnr lrndweyirf edrrnslpds sfdkvetppk
541 pikkdvrfak evclastwss nayeranpef imnrhrllwm mkvhpsmnsa mneiklelns
601 ykknemvvhe nskcfthyliBLASTp Search for Homologous Sequences
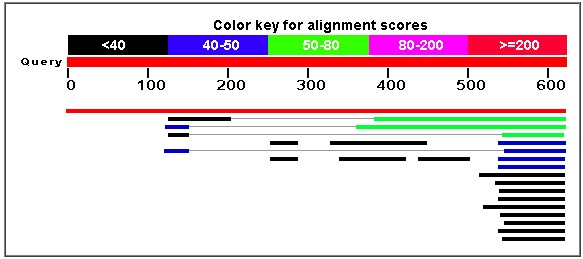

http://www.ncbi.nlm.nih.gov/blast/Blast.cgi
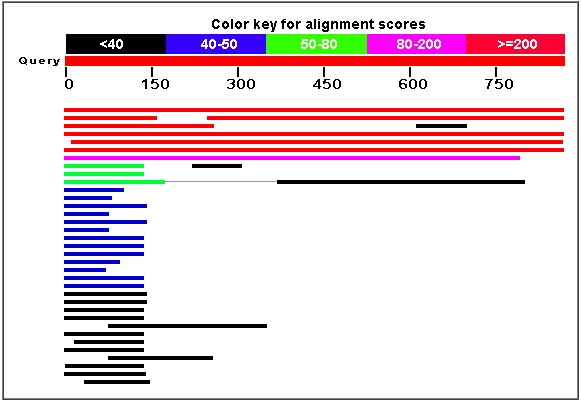
As one can see from the BLASTp results displayed in the screenshots above, the bit score and the E-value for the alpha-factor recepter regulator were the highest and lowest, respectively, by very considerable amounts. It is easy to say with certainty that amino acid sequence comes from ARF1.
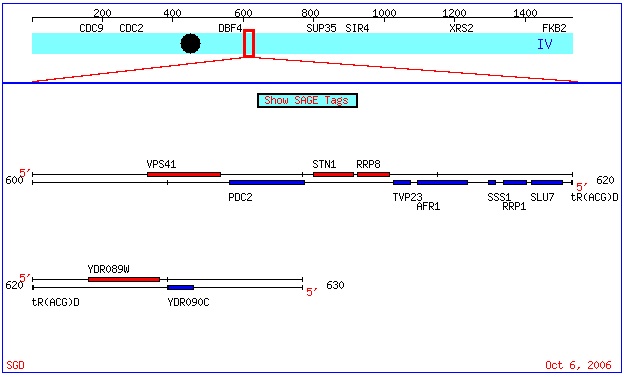
ORF Map
http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000002492
As one can see from the abve screenshot from the SGD site, AFR1 is Crick strand ORF, based upon its blue color. More information regarding the color code used is avaiable here.
Non-Annotated Gene: YDR089W
- Description -
The gene entitled Ydr089w is a non-annotated Watson strand ORF. The function and role of this gene is currently unknown.
Gene Ontology:
Molecular Function - unknown
Biological Process - possibly involved in membrane organizaion and biogenesis (based on sequence similarity) (Cohen, 1999; http://www.jbc.org/cgi/content/full/274/38/26885)
Cellular Location - cell membrane (based on sequence similarity) (Cohen, 1999; http://www.jbc.org/cgi/content/full/274/38/26885)
Location:
Chromosome IV, base pairs 616147 to 614285
- Amino Acid Sequence Information (from NCBI)
ORIGIN
001 mkfedrilnk sipewkfyni nyeklkvaik kvtaydydnp ndsgmeklln qcsvafdqef
061 qnvnlfvslk ikeistrils vessiidfsk glnktsrnrf nlrklkiina hvddcnfelq
121 llsrfliiqr ialrklfkkl lnefpqdsen pltaseyvts irnseslrng hegisfmkld
181 ldpyllevsl ivdvlhdlen kledatepav eqrslnrsde sahtssspea nnsslpaspr
241 sipllsnkkt skmidsslef dtalidkaen lgrfllssed ieglkfmlln igfriiddsi
301 istskeildt tdninsagnk sirsaksfnd lqhtlslskq knilpsavqs nekyvsisil
361 dtvgnegspl lltddninqh pnmivsstae dtcivmchvg glrnhvvtnd lllrdvknil
421 samrsgndtk nisalinsld pspiskiale wiqshrlkti epkldfkrtr fisadngdiy
481 lialdesiti gnvstlpfpi leikklsrss glsqtained nkfkqlmksv vtnefqcsli
541 ppdlttwkic lelvhsnelq ndlfqlllrd qyklnsddsl spdeffqlgk drleeefdlt
601 gpinnsqgsv dsgrrvrihk kskqsdnetk kkpirywnef deqeednldn afyidtngsr
661 sttdneesll lrnsppdygf ilfsrnfinr tydfceklrn lirhdkktsp dlfqnskhph
721 csstnygsva sfgsqstsas yddvqrylqy qqqdiedsqs iyeyrhdevv tflylsallt
781 scimasvclg ivlslfrgqs nneidleiqn iliaiiiisl lvslilicac llllfsrftl
841 apiwhyvgcf tmffsvtgtv cygmieiff
BLASTp Search for Homologous Sequences
http://www.ncbi.nlm.nih.gov/blast/Blast.cgiAs one can see from the above results produced by the BLASTp search, the best hit is definitely the protein we have in question, encoded by YDR089W. However, the next few hits are highly similar in sequence, with the second hit actually being 100% indentical. This would suggest that there is a perfectly homologous yet unknown protein produced by the unknown gene with the accession of CAA90448. Only a portion of the search results are displayed in the above figure, due primarily to the fact that the search returned a relatively large number of insignificant hits; only the top hits are included.
Further Homology Search
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=homologene&dopt=AlignmentScores&list_uids=38670
The above screenshot, taken from NCBI's HomoloGene search results for "YDR089W" shows the percent identity in both protein and DNA between YDR089W and two rather similar sequences for genes found in K. Lactis and E. gossypii. If you refer back to the BLASTp search results above, you will notice that these genes are included towards the top of the hits.
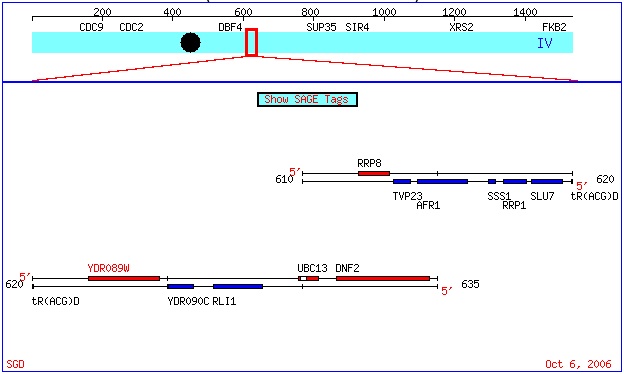
ORF Map
http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000002496
As one can see from the above screenshot from the SGD site, YDR089W is a Watson strand ORF, based upon its red color. More information regarding the color code used is available here.
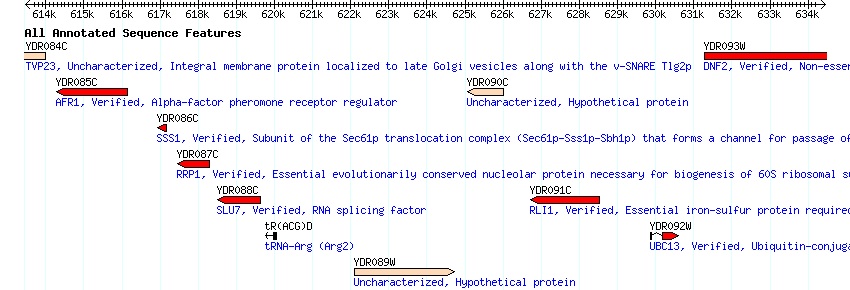
Predicted Secondary Structure
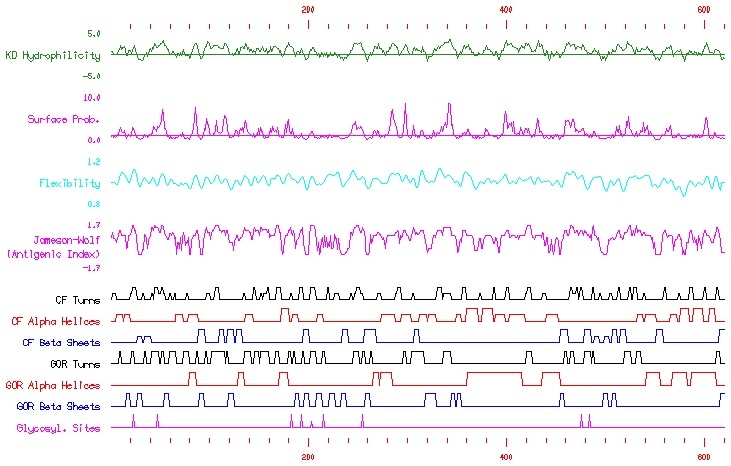
http://db.yeastgenome.org/cgi-bin/protein/secStructure?Type=PS&Feat=YDR085C
Looking at the above screenshot, taken from the SGD, one can notice where along the amino acid sequence turns, alpha helices, and beta shets are predicted to occur. Using this as a model allows for researchers to predict, although to lesser certainty, the tertiary structure of the protein.
Hypothesis: YDR089W is involved in the extension of the membrane during shmoo formation and alpha-factor reception.
References
- Bernd, Karen. "Cell Biology." Davidson College course notes, taught Fall 2004.
- Cohen, Adiel, et al. "A Novel Family of Yeast Chaperons Involved in the Distribution of V-ATPase and Other Membrane Proteins."Journal of Biological Chemistry, 1999. http://www.jbc.org/cgi/content/full/274/38/26885. Accessed 10/06/06.
- National Center for Biotechnology Information (NCBI). http://www.ncbi.nlm.nih.gov/. Accessed 10/06/06.
- Saccharomyces Genome Database (SGD). http://www.yeastgenome.org. Accessed 10/06/06. (All information from the two "Description" sections of this page was compiled from this site)
Links
Genomics
Page
My
Home Page