My Favorite Yeast Proteins
AFR1 & YDR089W
This web page was produced as an assignment for an undergraduate course
at Davidson College.
Building upon what's been learned about AFR1 and YDR089W, the examination of protein structure and interaction should allow for a greater hypothesis for YDR089W and possible inverse-expression relationship with AFR1.
AFR1
Structure:
Unfortunately, AFR1 has currently not been crystallized and images depicting its secondary and/or tertiary structure are not available. Searching the Protein Data Bank, ModBase, and PANTHER resulted in no successful matches. I was able to find, by simply utilizing the SGD site, that the transcribed protein has a molecular weight of 71,363 Da and is composed of 620 amino acids. Based soley upon the sequence of amino acids found within the protein, the following prediction was made as to its secondary strucure:

http://db.yeastgenome.org/cgi-bin/protein/secStructure?Type=PS&Feat=YDR085C
Interactions:
I decided to start my examination into the protein interactions of AFR1 by visiting both the Database of Interacting Proteins (DIP) and BioGRID. Surprisingly, the two databases returned distinctly different interactomes for the same protein. In the interactome below and to the left was obtained from DIP, and the 1st shell nodes (orange) represent the following, starting at the top and moving clockwise: BOI1, IQG1, SEN15, and CDC12. As one can easily see, these proteins are indeed represented on the interactome below and to the right, which was obtained from BioGRID. However, one notices the presence of three additional proteins: YRA1, GCN4, and STE2. DIP does not discuss any interaction with these three proteins in their site on AFR1, and hey are not represented in any of the twenty-five 2nd shell nodes (yellow). Inevitably this begs the question, why? Is one of the two interactomes outdated? Or, did DIP conclude that the evidence supporting the interaction between AFR1 and the three proteins in question was not strong enough to warrant inclusion in their interactome?
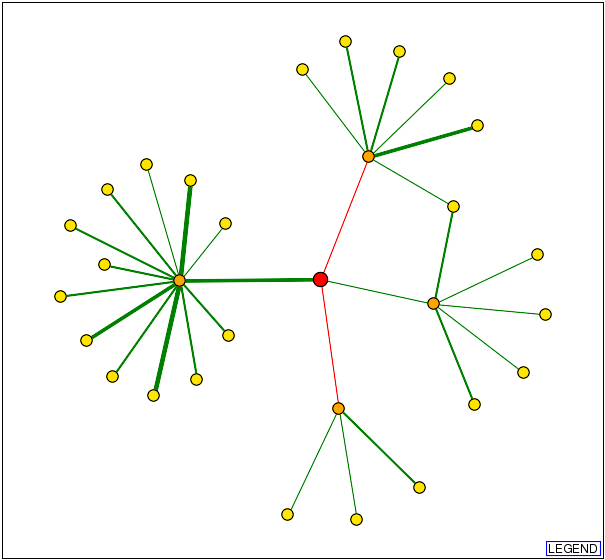
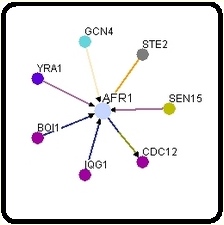
http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=824 http://www.thebiogrid.org/SearchResults/summary/32141
In a very small attempt to look into this matter a bit further, I examined where the evidence was coming from in BioGRID's construction of their interactome. As one can see from the table below the evidence for the YRA1, GCN4, and STE2 nodes appears to be valid, at least from a rather general glance. Although the following data does not answer the question of why the two interactomes differ, it does provide the reader with the evidence that BioGRID has used to compile theirs.
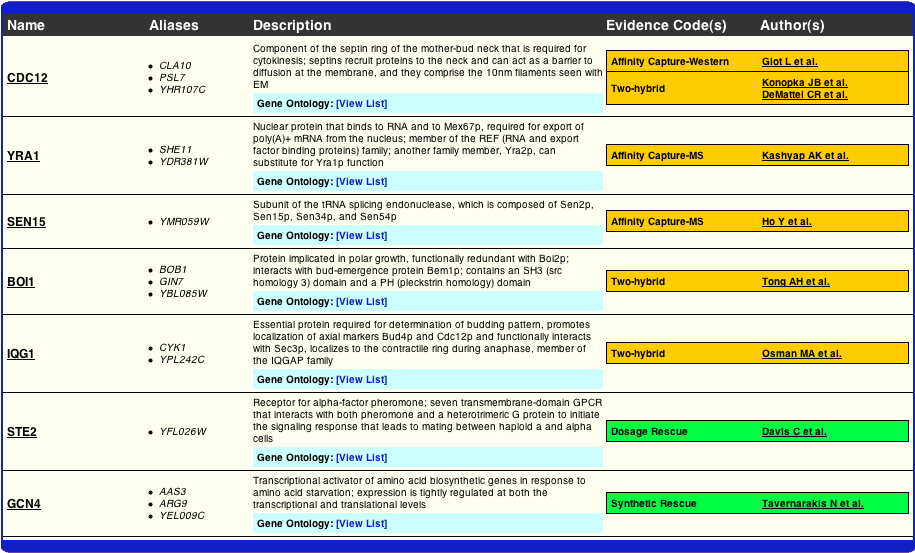
http://www.thebiogrid.org/SearchResults/summary/32141
Simply sticking to the four proteins that are included in both interactomes (BOI1, IQG1, SEN15, and CDC12), all appear to be involved either directly or indirectly in processes connected to alpha factor pheromone reception and regulation. Based on what we know and what I've presented in my previous pages, this makes perfect sense.
I would also like to note here, mainly for comparative purposes with YDR089W later on, that SGD predicted no shared domains, no unique domains, no transmembrane domains, and no signal peptides for AFR1.
YDR089W
Structure:
Unfortunately, YDR089W has currently not been crystallized and images depicting its secondary and/or tertiary structure are not available. Searching PDB, ModBase, and PANTHER resulted in no successful matches. I was able to find, using SGD, that the transcribed protein has a molecular weight of 98,711 Da and is composed of 869 amino acids. Based soley upon the sequence of amino acids found within the protein, the following prediction was made as to its secondary strucure:
http://db.yeastgenome.org/cgi-bin/protein/secStructure?Type=PS&Feat=YDR089W
Interactions:
Unfortunately, YDR089W has currently not been added to DIP or the BioGRID database, making direct data comparison with AFR1 rather difficult. However I was able to use the SGD to find that, in contrast with AFR1, nine shared domains were predicted for YDR089W. The following table represents those proteins which include the given shared domain, as well as other notable motifs found within the proteins.
The first thing that I noticed when examining this data was that all nine proteins share the exact same motif with YDR089W, that being SPX. Using this information, I used the Sanger Institute's Pfam database entry on SPX to explore the experimentally-observed function behind the SPX domain, which is summarized by the screenshot below.
http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF03105
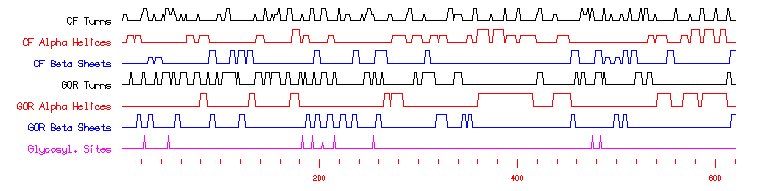
After having read this information, what obviously stands out the most is that all of the proteins that contain the SPX domain (including YDR089W by default) "are involved in G-protein associated signal transduction." Combing this knowledge with already knowing that YDR089W is a membrane protein of some sort allows for greater insight into the protein's possible role within the yeast cell. Further study of SGD's predictions on YDR089W showed that there are no predicted unique domains and no signal peptides. As one may guess, there were indeed predicted transmembrane domains - three to be exact. These three predicted domains occur from amino acids 776 to 798, 811 to 833, and 843 to 865. Simple math allows one to recognize that each predicted transmembrane domain is 22 amino acids long. While this may not enlighten us as to the function or interactions of the protein, it definitely hints more about the overall structure of the molecule. By comparing this information and the following graph with YDR089W's secondary structure prediction by SGD provided earlier, one can determine how the protein interacts conformationally with the plasma membrane (such as, appears as if these domains are dominated by CF Beta Sheets).
http://db.yeastgenome.org/cgi-bin/protein/domainPage.pl?dbid=S000002496#domains
Conclusions:
Although much was learned about YDR089W, there is still not enough information to support my previous hypothesis that it is a membrane protein involved in some aspect of sporulation. Where the first half of the hypothesis is supported, a lack of structural information and an interactome for the protein makes the determination of probable function difficult. Obviously, future experiments are needed in order to reacha better understanding.
Future Experiments
Obviously, my highest recommendation would be for the crystallization of both proteins. Such an accomplishment would allow for the determination of the secondary and tertiary structures. This would obviously facilitate further research involving the proteins, as well as allow for their eventual incorporation in the PDB. Ultimately it would be nice to have the proteins' structures viewable in Jmol format. If the proteins could be crystallized under various conditions (e.g. bound to substrates or interacting with the other aforementioned proteins) it would provide researchers with a more clear picture as to how exactly these proteins work.
A second experimental recommendation would be to try to see which proteins YDR089W interacts with. This could hypothetically be done via the crystallization mentioned above.
References
Links
Genomics
Page
My
Home Page