Abstract
Paraganglioma, also referred to as pheochromocytoma, is a rare form of cancer in which hormone-secreting tumors emerge on the head and neck (Bogdasarian 1979). Paragangliomas are highly variable in both aggressiveness and mobility and are characteristically difficult to treat, as conventional courses of cancer treatment are often ineffective (Jensen 1991). These inherent characteristics further contribute to the inability of clinicians to predict patient prognosis reliably, although recent corroboration of a clear a link between increased malignancy and mutations in the succinate dehydrogenase subunit B (SDHB) gene hold promise for improving diagnostic resolution (Bogdasarian 1979, Jensen 1991, Smith 2007). Initial studies in this vein have shown that succinate accumulation, resulting from the SDHB mutation, prevents degradation of the known oncogenic transcription factor, HIF1α (Qutub 2007). Additional labs have contributed to extensive research in paraganglioma models containing the SDHB mutation, but the literature has yet to report specific mechanisms behind the unusually variable malignancy in paraganglioma (Boedeker 2007, Baysal 2007). As part of my BIO309 course investigation, I explored the human SDHB gene, using publicly available genomic and proteomic tools, in search of genetic anomalies that might contribute to differential malignancy in paraganglioma. My research identified a SNP-containing MZF1 binding domain upstream from the SDHB gene (Ensembl, BLAST, Jaspar, Morris 1994). MZF1, a known tumor suppressing factor, is a critical mediator of cell proliferation and migration (Gaboli 2001, Hsieh 2007). I propose a putative mechanism by which misregulation of SDHB by MZF1 contributes to increased malignancy via disinhibition of HIF1α signal. Furthermore, I provide a model of the hypothesized interactions to identify potential therapeutic targets. If validated, these findings would greatly improve clinicians’ ability to both diagnose and treat paraganglioma.
Introduction to Paraganglioma: From Clinical Limitations to Laboratory Research
The Problem : Paraganglioma manifests in patients with tumors that vary in aggressiveness and mobility, which corresponds to highly variable patient prognoses. The most aggressive tumors reported in the literature are highly malignant, and are essentially untreatable. Our current understanding of the increased risk for malignancy in paragangliomas is inadequate. As a result, patients who present with tumors have very few options for treatment.
Why Study Paragangliomas? The tumors of a host of other cancers are characteristically malignant. Is there some reason why we should focus so specifically on paragangliomas, given their relative rarity?
- Paragangliomas draw the interest of researchers primary because of their particularly significant impact on patients' quality of life.
Paragangliomas belong in a particular class of tumors known as Neuroendocrine Neoplasias. The name refers to the ability of these tumors to overproduce catecholimine neurotransmitters (e.g. dopamine and norepinephrine). This subsequent secretion of these neurotransmitters into the circulatory system initiates a devastating system-wide response that can completely incapacitate affected individuals.
How is Paraganglioma Currently Treated?
Surgical Excision: Surgical intervention is often the best treatment option for patients, although the success rate is still very low (Chrisoulidou 2007).
-Paragangliomas have a high remission rate (Askainen et al 2005). Patients often have no choice but to undergo repeated surgeries.
-Satellite tumors are much more likely to gain dominance after excision of the primary tumor than is the case in other types of cancer. This characteristic is putatively attributed to the unique role played by HIF1α in these tumors (Emara 2007).
Chemotherapy: Chemotherapy is often used, but rarely offers any curative power to paraganglioma patients (Chrisoulidou 2007).
What Needs to Happen Next?
The primary goal of current research is to identify biological markers that can be used to both develop treatments and enhance the diagnostic capacity of clinicians. So far, researchers have been unsuccesful in identifying effective drug targets, but our understanding of how the disease comes about is improving. Most significantly, researchers have identified a potent transcription factor, HIF1α, as the primary contributor to tumirogenesis in paragangliomas. Furthermore, succinate accumulation due to mutations of the SDH complex have been identified as the mediating factor involved in the overexpression of HIF1α. The next step requires further characterization of the driving circuitry behind pathogenesis, with the ultimate goal of tailoring drug treatments.
My Goal: To use publicly-available genomic tools to investigate possible contributions of gene misregulation in paraganglioma malignancy.
The SDHB gene: Implicating Gene Misregulation
The SDHB gene has been highly conserved throughout evolution, with its closest current relatives in Pan troglodytes and Mus musculus (ClustalW). Despite the similarity of their respective orthologs, P. troglodytes and M. musculus do not display the human disease phenotype upon SDHB mutation (Piruat 2004). As a result, paraganglioma researchers favor simpler, more cost-efficient animal models to study the genomic and proteomic interactions that underlie the disease (Dow 2007). So, research of paraganglioma symptomology is restricted to clinical observation and case study, leaving small organism research with the responsibility of corroborating, and better explaining the observed correlation between mutated SDHB and malignant paraganglioma. Current research has done well to identify several mutations within the SDHB might give rise to the paraganglioma phenotype, however analysis of these mutations has failed to account for the unusual variability of malignancy in these tumors.
HYPOTHESIS: I predict that misregulation of SDHB expression in paraganglioma tumor cells contributes to variable malignancy via exacerbation or attenuation of succinate accumulation.
REASONING: Researchers have implicated gene misregulation in the pathogenesis of many forms of cancer (Heim 1991, Saporita 2007). The dynamic transcriptome of the SDHB gene responds to changing cellular environments through the complex interaction of a variety of transcription factors (Neumann 2004). If this regulatory machinery fails to properly regulate SDHB expression in the stressful conditions in tumor cells, a further reduction of SDHB function could occur, effectively hastening tumorigenesis and/or malignancy in a given individual.
Important Consideration: Reduction vs. Loss of function
The first issue I encountered in developing my hypothesis was that current reports have suggested that familial paragangliomas are typically transmitted by SDHB mutations that correspond to a complete loss of SDHB function (Bogdasarian 1979, Jensen 1991, Smith 2007). In order for my investigation of SDHB misregulation in paragangliomas to be valid, I first had to establish the potential for SDHB mutations that corresponded to a reduction in function as opposed to a complete loss thereof.
I then considered the three iron-sulfur clusters in the SDHB protein that ultimately confer the electron-transporting capacity of SDHB (Smith 2007). Evaluation of predicted hydrophobicity and/or hydrophilicity of the protein, using Kyte-Dolittle and Hopp-Woods analyses (window = 7), respecively, failed to provide any indication of where the functional clusters were located. I decided that I would have to turn to other sources for more useful information. To guide my search, I postulated that small mutations in one or more, but not all of these functional units could confer a reduction in biological function without complete inhibition. As noted, paraganglioma mutations are conventionally thought of as corresponding with complete loss of function, however, there is emerging evidence in favor of my predictions (Cheng 2004, Lima 2007).
Based on these prelimiary findings, I established a subjective set of criteria by which I determined the validity of investigating SDHB misregulation: In order to feel compfortable proceeding, I wanted to locate candidate mutations in biologically significant regions of the SDHB gene, and more specifically in those sequences that correspond to iron-sulfur domains.
Determining Biologically Significant Regions of the SDHB gene:
I began determining the focus of my search according to the principle that the most important, or functionally-significant, portions of a given gene are the least likely to change over time (Hawkins 1991). In keeping with this evolutionary phenomenon, I turned to NCBI's Conserved Domain Database and searched the amino acid sequence of theSDHB protein for its most highly conserved regions (NCBI-CDD). The database identified a large, highly conserved domain, sdhB (E = 8 * 10-109), which occupies the portion of the SDHB protein from amino acid 40 to 270 (Figure 1(a)) .
I then stepped back to consider the entire SDHB so I could determine if there were any iron-sulfur binding features that occured in or overlapped with the sdhB domain. Usingly publicly-available genomics resources, I noted the presence of the ferredoxin feature over the range of a.a. 40 to 133 on SDHB (InterPro) (Figure 1(b)). This feature contains the [2Fe-2S] iron-sulfur clusters of the SDHB protein, making the feature ideal for my search (Smith 2007). I began turning the focus of my search to the genetic sequence encoding these features, and converted all necessary amino acid coordinates to base pairs to determine location of key portions of the sdhb gene.
| Figure 1: Results from the NCBI Conserved Domain Database Search. | |
| (a) Note that the sdhB CD is the most highly conserved, with an e-value of 8e-109. This domain provided the initial focus for my search. | 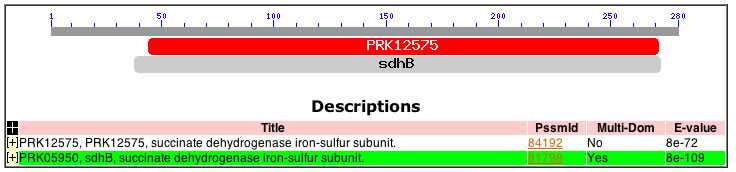 |
| (b)Representation of InterPro domain architechture results. Note the presence of the ferrodoxin domain on the reverse strand within the sdhB CD of the sdhb gene. | 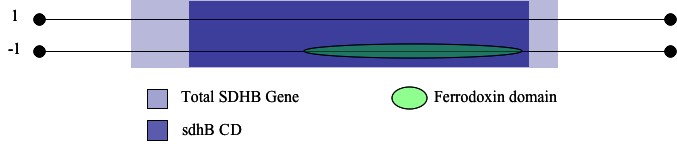 |
Locating Single Nucleotide Polymorphisms (SNPs):
I chose to search for potential disease mutations by locating any known SNPs in a biologically significant portion of the SDHB gene (identified above). I restricted my search of the SDHB-coding transcript by calculating the range of base pairs that correspond to the range of the SDHB protein occupied by sdhB. I then identified six potentially-disease causing SNPs contained in the specified region (Ensembl, Figure 2). Five of the six were nonsynonymous coding SNPs which occurred at amino acids 53, 57, 60, 119, and 163. The sixth represented a frameshift coding SNP that occured at a.a. 254. I considered each of these snps to be a candidate for a reduction of function mutation. I became even more optimistic when I determined that the sequence corresponding to the ferredoxin feature contained four of the six SNPs identified in this analysis (Figure 2).
| Figure 2: Location of SNPs within the sdhB conserved domain and the ferrodoxin feature. |
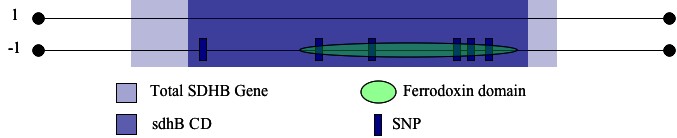 |
| Note the numerous SNPs within the ferrodoxin domain. Due to the role played by the 2Fe2S clusters within this domain, single nucleotide alterations could hinder the ability of the subunit to perform proper electron transfer. |
Looking Upstream
Having identified several putative reduction of function mutations, I turned my attention to the 500 base pairs immediately upstream from SDHB. I recognized that in order to maintain an ability to infer potential implications for a given SNP in this sequence, I would need to know something about the regulatory interaction it would hypothetically affect. With this in mind, I searched the upstream sequence for known DNA-binding motifs and determined that the sequence contains several putative transcription factor binding sites (PREDATOR). I did not want to characterize the nature of each possible hit just yet, however, as the motifs would not mean much in the way misregulation unless there was a potential for the nature of that site to be altered in some way. Again, I turned to SNP identification as an effective way of searching for potential mutations in the region. Of the motifs identified in my initial search of the upstream sequence only one, the MZF1 binding site (score = 8.89), contained a potentially disease-causing SNP (Figure 3). The SNP occurs 293 base pairs upstream from SDHB and corresponds to a G/A switch. In this portion of my analysis I had come upon what I set out to find, a genetic basis for potential misregulation of SDHB expression.
| Figure 3: Graphical summary of SNP-containing MZF1 binding motif [Ensembl, JASPAR] |
 |
| Note that the motif and SNP are located immediately upstream (red shaded region represents 500 base pairs) from the sdhb gene. |
MZF1, or myeloid zinc finger 1, is well documented as a powerful regulator of cell proliferation, adhesion, mobility (Gaboli, 2001). More importantly, MZF1 is a known tumor supressor gene (Gaboli, 2001). The activity of the transcription factor on mitochondrial genes is not well characterized, although it presents itself as an ideal candidate for the present analysis.
REFERENCES
- Bogdasarian RS and Lotz PR: Multiple simultaneous paragangliomas of the head and neck in association with multiple retroperitoneal pheochromocytomas. Otalaryng Head Neck Surg 1979, 87: 648-652.
- EBI Tools: ClustalW [http://www.ebi.ac.uk/Tools/clustalw/index.html]
- Ensembl Genome Browser [http://www.ensembl.org/index.html]
- Hawkins AJS: Protein turnover: A Functional Appraisal. Functional Ecology 1991, 5(2):222-233.
- Heim K, et al: Selective repression of retinoic acid target genes by RIP140 during induced tumor cell differentiation of pluripotent human embryonal carcinoma cells. Mol Cancer 2007, 6(1):57.
- Jensen JC, et al: A report of familial carotid body tumors and multiple extra-adrenal pheochromocytomas. J Urol 1991, 145:1040-1042.
- Kyte-Dolittle Hydropathy Plot [http://gcat.davidson.edu/rakarnik/kyte-doolittle.htm]InterPro - http://www.ebi.ac.uk/interpro/IEntry?ac=IPR001041
- Li FP: Identification and Management of Inhereted Cancer Susceptibility. Environmental Health Perspectives 1995, Suppl 8 (103):297-300.
- Lima, J, et al: Mitochondrial succinate dehydrogenase structure-function relationships and clinical-pathological correlations. J Clin Endocrinol Metab 2007, No. 17848412.
- Manton KG, Lin K: Projecting Chronic Disease Prevalence. Medical Care 1984, 22(6):511-526.
- Myers ER, et al: Genomic tests for ovarian cancer detection and management. Evid Rep Technol Assess 2006, 145:1-100.
- NCBI Conserved Domain Database [http://130.14.29.110/Structure/cdd/cdd.shtml]
- Neumann HPH, et al: Distinct clinical features of paraganglioma syndromes associated with SDHB and SDHD gene mutations. JAMA 2004, 292: 943-951.
- Piruat JI, Pintado CO, Ortega-Saenz P, et al: The mitochondrial SDHD gene is required for early embryogenesis, and its
- PREDATOR Secondary Structure Prediction Method [http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html]
- Saporita AJ, et al: Therapeutic targets in the ARF tumor suppressor pathway. Curr Med Chem 2007, 14(17):1815-27.
- Smith EH, Janknecht R, Maher LJ: Succinate inhibition of {alpha}-Ketoglutarate-Dependent Enzymes in a Yeast Model of
- Paraganglioma. Hum Mol Genet 2007, [Epub ahead of print].
- The Jaspar Database [http://jaspar.cgb.ki.se/]
Exploring the Role of Gene Misregulation
Key Concepts:
- Mutations in SDHB lead to tumorigenesis by triggering HIF1α hypoxia signal via succinate accumulation (Smith, 2007; Guzy, 2007).
- The rate of mutagenesis is dependent on the intensity of positive feedback loop where HIF1α represses SDHB expression, leading to a further succinate buildup, which further disinhibits HIF1α production and signaling.
What do we know? Tumors produce a wide range of environmental stressors beyond HIF1α. This phenomenon corresponds with the activation of any number of stress signaling pathways.
MY QUESTION : Can inhibition of MZF1 activity or other transcription factors on SDHB exacerbate the positive HIF1α feedback loop?
TURNING TO A COMMONLY USED MODEL EUKARYOTE: Saccharomyces cerevisiae
-When using an animal model in research, the most important consideration is whether or not the corresponding ortholog for your gene of interest is similar enough to allow for inferences to be drawn about the human gene's function?
In this case: Is the yeast gene, SDH2, close enough to SDHB to justify its use in studying the biochemical correlates for paraganglioma pathogenesis?
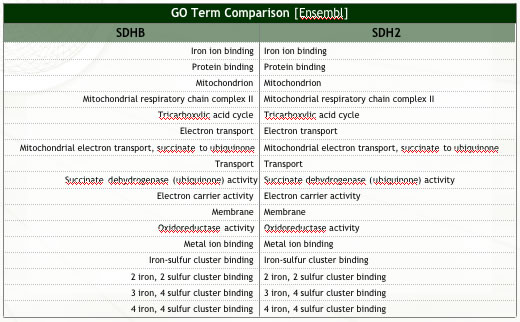
- I performed a number of comparisons (Figure 1) and found that the answer is YES, SDH2 function is highly relevant to that of SDHB. This similarity in biological function overrides the somewhat lacking similarity in other areas (refer to Figure 1). In fact, S. cerevisiae mitochondrial genes are often used to model human diseases due to their functional relatedness (Steinmetz, 2002).
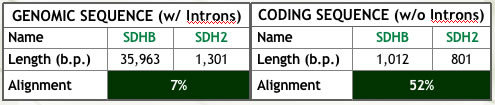
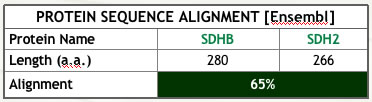
| Figure 1: Summary of analyses conducted to assess ortholog similarity. | |
(a) Alignment of the genomic and coding sequences (Ensembl). Note the clear effect of intron content on similarity. |
|
 |
|
| (b) Protein sequence Alignment (Ensembl, JalView). The proteins are of similar length, although their alignment is far from ideal | |
 |
 |
| (c) Ortholog functional similarity (Ensembl). Note the two orthologs perform identical (known) functions. | |
 |
|
The Hoopes' Lab Experiments: Microarray Data Analysis
MSN2/4 Mutation:
- Msn2/4 are zinc-finger binding proteins (recall MZF1) that mediate a wide variety of cellular stress responses (Gasch, 2000). The following outline briefly summarizes some of the known Msn2/4-mediated stress signals:
- Oxidative Stress (Martinez-Pastor, 1996)
- Age-dependent response to oxidative stress; increased risk of cancer in elderly (Fabrizio, 2001).
- Hindrance of stress response mechanisms over time has been implicated in the increased likelihood of cancer as age increases (Saunders, 2007).
- The transcription factors are found in the nucleus and in the cytosol (Gorner, 1998; Mayordomol, 2002).
NOTE: There is no mzf1 ortholog in yeast. As such, all subsequent analyses were designed to focus more generally on regulation of SDHB expression in an organism whose stress stress response machinery is damaged.
Question #1: How is SDH2 regulated in the absence of proper Msn2/4 function? Will regulation of SDH2 be different during the shift from log phase to stationary phase?
- To determine the answer I used a two column plot comparing expression of the entire genome during the shift from log to stationary phase with or without Msn2/4. I present the results of this analysis below (Figure 2).
| Figure 2: Analysis of Microarray Data from Hoopes' Laboratory Experiments. MSN mutant data is plotted along the y-axis and wild type along the x-axis (Dissimilarity threshold = 0.1). Mitochondrial Respiratory Chain Complex II Genes are plotted in red. |
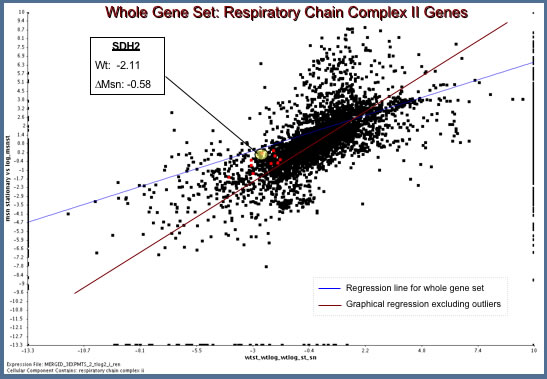 |
The Hoopes' Lab data reveal an interesting result.
- In wild type S. cerevisiae, SDH2 is down-regulated (Ratio: -2.11) during the transition from log to stationary phase.
- In the msn2/4 mutated strain, however, there is a failure to downregulate SDH2 expression.
- What implications do these findings have in SDHB regulation of paragangliomas?
These findings offer evidence in support of the notion that SDHB expression is regulated by zinc finger transcription factors. The data do not allow us to make more bold inferences.
Limitations of the Hoopes' Study (besides the dissimilarity between MZF1 and Msn2/4):
- We cannot conclusively say why there was no change in SDH2 expression in msn2/4 mutants. It could be that SDH2 was nont down-regulated in stationary phase, or that it was not up-regulated in log phase. We can suggest that the most likely possibility is the latter, but we are not afforded any capacity to conclusively prove it by the data.
To reiterate: We can say, based on these data, that damaged cell machinery related to stress response does affect SDH2 regulation.
TURNING TO THE LITERATURE: What Evidence can we find that supports upregulation of SDH2 in stress conditions?
- Stress condition: Glucose Limitation (Ferea et al, 1999)
- Three long term chemostats were inoculated with a single parental diploid strain. The parental strain was reared in normal YPD media, the three evolutionary strains were reared in glucose-deprived media.
Finding: SDH2 expression was clearly upregulated in response to stress via glucose-deprivation.
- Stress condition: Heat Shock (Gasch et al, 2000)
- Transcription of the entire S. cerevisiae genome was quantified as cultures were subjected to a variety of environmental changes. Of the most relevance to the current study was the induction of a generalized stress response via heat shock (cultures reared in either 25°C or 30°). Ten samples were collected over a period of 5 days and genome expression was assessed at each interval.
Finding: SDH2 was upregulated dramatically, peaking at 49.9 fold induction.
What are the implications of these studies?
- Recall: Paraganglioma pathogenesis arises and is driven by overexpression of HIF1α, which reinforces its own signal by downregulating SDHB expression in humans.
- Due to the lack of an MZF1 ortholog, I set out to analyze yeast microarray data in search of any evidence suggesting an induction of SDH2 (SDHB's ortholog) during stress conditions.
- While the Hoopes' Lab data was somewhat dissappointing in its relevance, a literature search revealed a well established activation of SDH2 during stress in S. cerevisiae.
TRIPLES Database Search (Kumar et al, 2000; Ross-Macdonald et al, 1999): Have any mTn mutants been created in the SDH2 gene in yeast?
The mTn mutations provided by the TRIPLES Database revealed little utility for future study of SDH2. There were four phenotypes induced by transposon insertion, but three of them grew just as well as wild-type strains in 20 µg/µl Benomyl medium (an anti-fungal agent). The fourth strain grew at a 'medium' rate relative to wildlife in a haploid viability test, which offers little exerimental utility. There were a total of six clones produced (4 insertions out-of-frame, 2 in-frame), yet the disruption by insertions in each case was minimal. All insertions in all clones had sense LacZ orientation. No noteworthy results in the gene expression data either.
Attempting to study SDHB regulatory interactions in yeast is clearly difficult, and I have not had tremendous success in my analysis thus far. One fact is self-evident: We need to learn more about SDH2 in yeast in as much relevance as possible for paraganglioma pathogenesis.
PROPOSED FUTURE STUDY: Focusing on regulation by zinc finger proteins
Recall that HIF1α signaling is directly related to induction and maintenance of cellular response to hypoxia. I propose a study in which regulation of SDH2 is challenged by mutating all known zinc finger binding domains within 1000 base pairs upstream of SDH2. The goal of this manipulation is to determine the effect of loss of zinc finger regulation on SDH2 during paraganglioma-type hypoxic condition. The design follows (Figure 3):
Figure 3: Proposed Future Study |
||
| (a) Experimental Setup | ||
 |
||
| (b) Manipulation: Stressor Administration | ||
 |
||
| (c) Expectations | ||
| IF zinc finger regulation is important in response of SDH2 to hypoxia conditions: | IF zinc finger regulation is NOT important in response of SDH2 to hypoxia conditions: | |
 |
 |
|
(d) MICROARRAY SETUP
|
||
Design |
Reasoning |
|
| Platform | PCR-amplified cDNA for all genes in the yeast genome SDH2 cDNA |
To allow for analysis of a wide variety of expression profiles which could lead to identification of previously uncharacterized coregulation patterns. |
| Positive Control | Blank spots | To verify detection of gene of interest, which should be produced in both experimental group. |
Negative Control |
cDNA for GFP gene | Nothing should be detected at either spot. -Blank: no cDNA -GFP: not produced |
REFERENCES
Bogdasarian RS and Lotz PR: Multiple simultaneous paragangliomas of the head and neck in association with multiple retroperitoneal pheochromocytomas. Otalaryng Head Neck Surg 1979, 87: 648-652.
EBI Tools: ClustalW [http://www.ebi.ac.uk/Tools/clustalw/index.html]
Ensembl Genome Browser [http://www.ensembl.org]
Fabrizio P, et al: Regulation of longetvity and stress resistance by Sch9 in yeast. Science 2001, 292(5515):288-90.
Ferea TL, et al: Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci USA 1999, 96(17):9721-6.
Gasch AP, et al: Genomic expression programs in the respone of yeast cells to environmental changes. Mol Cell Bio 11 (12):4241-57.
Gorner W, et al: Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev 1998, 12(4):586-97.
Guzy RD, et al: Loss of SdhB, but not SdhA, subunit of Complx II triggers ROS-dependent HIF activation and tumirogenesis. Mol Cell Biol 2007, [Epub ahead of print]
Hirama M, et al: Characterization of mitochondria in cisplatin-resistant human ovarian carcinoma cells. Oncol Rep 2006, 16(5):997-1002.
Jensen JC, et al: A report of familial carotid body tumors and multiple extra-adrenal pheochromocytomas. J Urol 1991, 145:1040-1042.
Kumar A, et al: TRIPLES: a Database of Gene Function in S. cerevisiae. Nucleic Acids Res 2000, 28:81-84.
Li FP: Identification and Management of Inhereted Cancer Susceptibility. Environmental Health Perspectives 1995, Suppl 8 (103):297-300.
Liu J, et al: Cadmium induced MTs synthesis via oxidative stress in yeast. Mol Cell Bio 2005, [Online]:139-145.
Manton KG, Lin K: Projecting Chronic Disease Prevalence. Medical Care 1984, 22(6):511-526.
Martinez-Pastor MT, et al: The S. cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element. Embo J 1996, 15(9):2227-35.
Mayordomo I, et al: Convergence of the target of rapamycin and the Suf1 protein kinase pathways in the regulation of subcellular localization of Msn2, a transcriptional activator of STRE-regulated genes. J Biol Chem 2002, 277(38):35650-6.
Myers ER, et al: Genomic tests for ovarian cancer detection and management. Evid Rep Technol Assess 2006, 145:1-100.
Ross-Macdonald, et al: Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 1999, 402: 413-418.
Saunders LR, Verdin E: Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene 2007, 26(37):5489-504.
Smith EH, Janknecht R, Maher LJ: Succinate inhibition of {alpha}-Ketoglutarate-Dependent Enzymes in a Yeast Model of Paraganglioma. Hum Mol Genet 2007, [Epub ahead of print].
Steinmetz LM, Scharfe C, et al: Systemic screen for human disease genes in yeast. Nat Gen 2002, 31: 400-404.
The Jaspar Database [http://jaspar.cgb.ki.se/]
Paraganglioma Disease Model: Genomic Strategy for Improved Treatment and Diagnosis
So far, we have been collecting pieces of a puzzle and now it's time to put them all together. I have synthesized the findings of my analysis into a model of paraganlioma in which I posit a putative mechanism by which hindered MZF1 activity on SDHB leads to increased malignancy of the associated paraganglioma (Figure 1- click to enlarge images).
| Figure 1 : Proposed interaction model for paraganglioma pathogenesis. | |
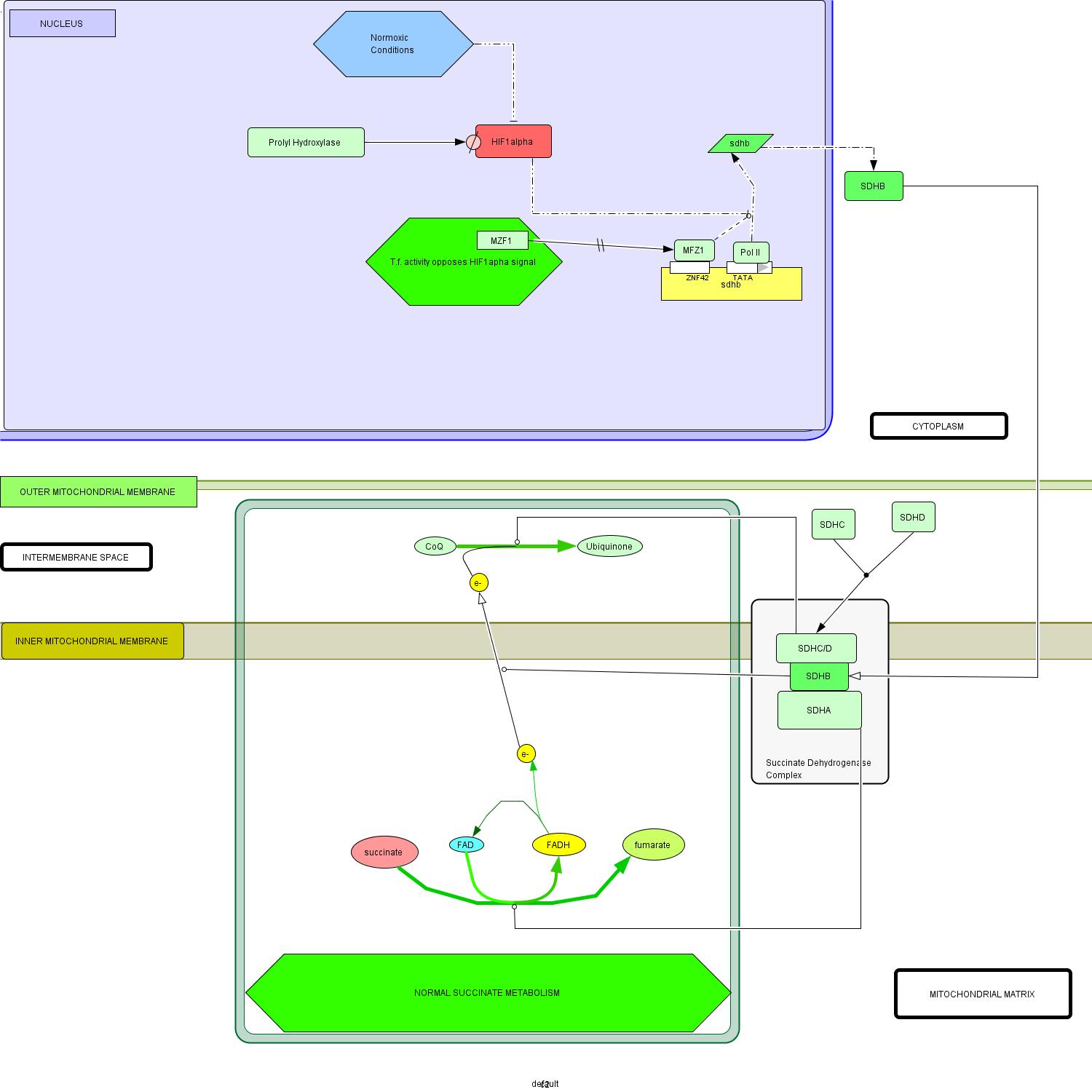
(a) Schematic representation of normal succinate metabolism. The function of SDHB is represented as a catalysis of electron transport from the mitochondrial matrix to the inter membrane space. NOTE: HIF1 alpha signal is suppressed through trnascriptional inhibition and enzymatic degredation, by prolyl hydrases. |
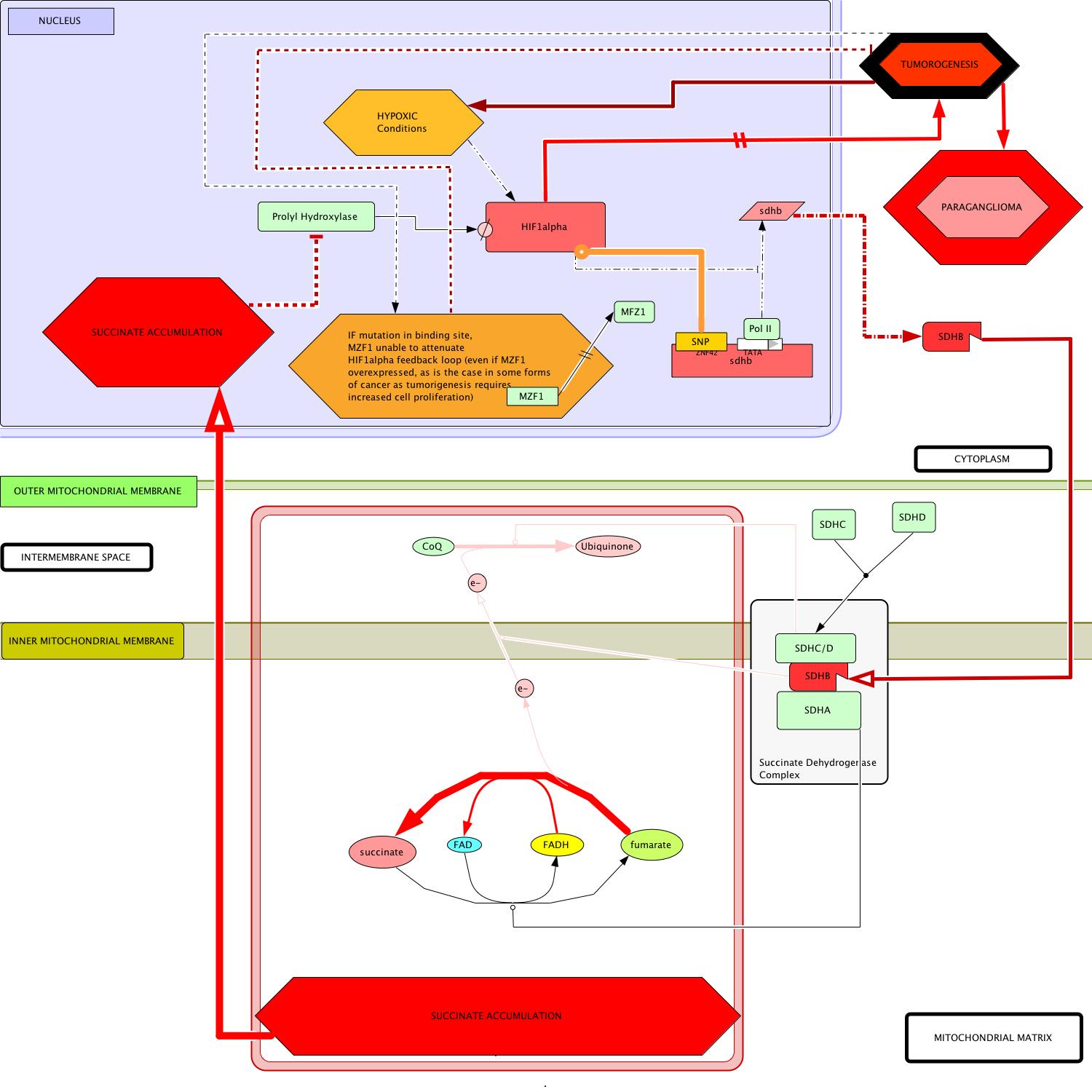
(b) Schematic representation of key changes in metabolic circuitry that result from mutations in SDHB. Note that succinate disinhibits HIF1 alpha by preventing its degredation. HIF1 alpha is overexpressed in the developing hypoxic environment and initiates its own positive feedback by transcriptionally inhibiting SDHB expression. If unable to bind to the SDHB gene, MZF1 cannot attenuate HIF1 alpha's effect on SDHB expression. This removal of inhibition will likely contribute to a drastic increase in the time-to-onset for the disease and malignancy of the presumptive tumor cells. |
Wild-type Model: Figure 1(a)
[Note: This model is simplified, avoiding discussion of the detailed mechanisms by which some processes occur. This is done to facilitate a more direct discussion of the key aspects of the circuit most relavent to paraganglioma pathogenesis.]
- Start: Normal transcription of the sdhb gene is mediated by RNA polymerase II and a variety of other regulatory factors, including MZF1 (Gaboli et al, 2001.
- Translation of sdhb mRNA occurs in the cytoplasm whereupon the SDHB protein is transported to its ultimate location in the succinate dehydrogenase complex (SDH), with its specific localization in the portion of the complex oriented toward the mitochondrial matrix (Pawlu et al, 2005).
- The remainder of the complex is formed by the addition of a heterodimerized transmembrane complex of SDHA/SDHB and the hydrophilic mitochondrial matrix-exposed SDHA subunit (Oyedotun and Lemire, 2004).
- SDHA catalyzes the conversion of succinate to fumarate, a reaction which transfers an electron to the FADH carrier (Bayasal et al, 2007; Briere et al, 2006).
- SDHB is a key mediator of directing electron flow (through its iron-sulfur clusters) across the inner mitochondrial membrane to the periplasmic space (Cheng et al, 2006).
- The SDHC/SDHD complex catalyzes the subsequent oxidization of CoQ to Ubiquinone, via the electron provided by SDHB activity (Doring et al, 2007).
Environment in the nucleus:
- In the normal state, MZF1 provides activating regulatory influence on transcription of the sdhb gene (Gaboli et al, 2001).
- Under normoxic conditions, prolyl hydroxylase enzymes actively degrade HIF1α (Asikainen et al, 2005, Frede et al, 2007).
- Thus, in addition to reduced transcription activation of HIF1α mRNA, the otherwise repressive activity of the hypoxia-inducible factor on sdhb transcription is extinct in normoxic conditions (Woodward and Maher, 2006).
Paraganglioma Disease Model : Figure 1(b)
[Note: This model is simplified, avoiding discussion of the detailed mechanisms by which some processes occur. This is done to facilitate a more direct discussion of the key aspects of the circuit most relavent to paraganglioma pathogenesis.]
- Start: In cases of reduction of function mutation (previously discussed), transcription of the altered sdhb gene produces a hindered protein product (Amar et al, 2007; Boedeker, 2007; Mannelli et al, 2007).
- Translation of mutant sdhb mRNA occurs in the cytoplasm whereupon the malfunctioning protein is transported to its same location in the succinate dehydrogenase complex (SDH).
- The remainder of the complex is formed as it normally is, by the addition of a heterodimerized transmembrane complex of SDHA/SDHB and the hydrophilic mitochondrial matrix-exposed SDHA subunit (Oyedotun and Lemire, 2004).
- In this disease model, SDHA still catalyzes the conversion of succinate to fumarate, producing the FADH carrier (Bayasal et al, 2007; Briere et al, 2006).
- KEY DEVIATION: The malfunctioning SDHB is now less able to direct electron flow across the inner mitochondrial membrane to the periplasmic space.
- Downstream in the circuit, the catalytic activity of SDHC/SDHD becomes all but irrelevant in the absence of the provision of electrons provided by a functional SDHB protein.
- Without proper direction by SDHB, the electrons on FADH remain localized in the mitochondrial matrix are used in the evergetically-favored reversion of fumarate back to succinate.
- Succinate accumulates in the mitochondrial matrix and slowly diffuses to the nucleus, where its buildup indirectly induces tumorigenesis (Pollard et al, 2005).
Environment in the nucleus:
- In the nucleus, succininates direct effect is inhibition of prolyl hydroxylase activity which, in effect, disinhibits the HIF1α signal (Ema et al, 1999).
- This disinhibition initiates a cascade of activity that becomes a positive reinforcement loop of the HIF1α signal.
- HIF1α overexpression directly contributes to the onset and proliferation of tumirogenesis (Cairns et al, 2007).
- Furthermore HIF1α represses the expression of the malfunctioning sdhb gene which subsequently exacerbates succinate accumulation in the cell, further disinhibiting the HIF1α signal.
- As the tumor grows, dying cells from the interior initiate natural HIF1α hypoxia signals which adds yet another reinforcing influence to the HIF1α loop.
- MZF1-mediated activation of transcription of the sdhb gene, suggests itself as an important mitigating factor against the positive reinforcement of the oncogenic HIF1α signal.
- Therefore, if the integrity of this opposing influence is compromised (as by a SNP in the t.f.'s binding site on the SDHB promoter) the associated paraganglioma will gain increased malignancy and present a greater challenge to currently utilized cancer treatments.
PROPOSING A GENOMIC STRATEGY FOR IMPROVED THERAPY FOR AGGRESSIVE PARAGANGLIOMA:
As I became aware in developing my model for paraganglioma pathogenesis, the accumulation of succinate due to SDHB mutations is a key element in the circuit promoting aggressive malignancy through disinhibition of the HIF1α signal. I propose the use of RNA antiswitch technology to use improper succinate accumulation as a trigger for inhibition of HIF1α transcription (Figure 2).
Other key elements of the design follow Figure 2.
| Figure 2: Proposed Therapy for Malignant Paraganglioma. Proposed antiswitch indicated in black, yellow 'S' ovals indicate succinate accumulation in the nucleus, and the triangle projection/indentation on HIF1α, antiswitch, and HIF1α-specific t.f. represents optimal target for selective inhibition of HIF1α transcription. |
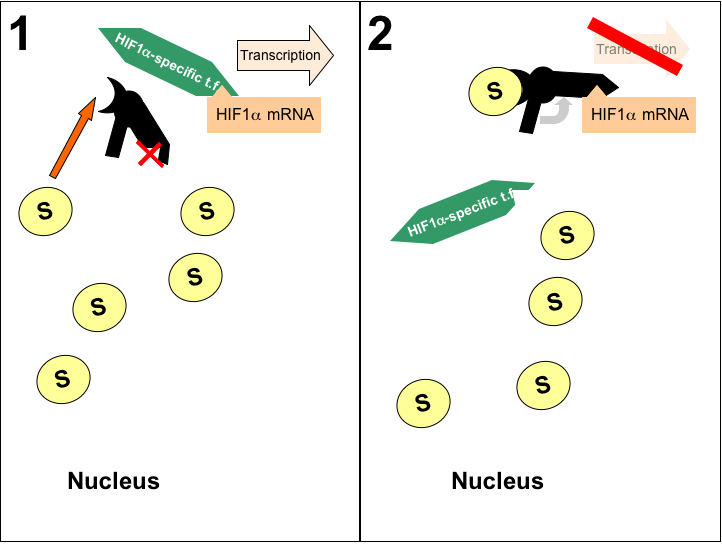 |
A. Determining the optimum target:
Successful implimentation of this therapy requires a target on the HIF1α promoter that is specific to that molecule. The Bioinformatics Research Group provides a listing of several known antisense RNAs for HIF1α. These RNAs provide a useful starting point from which the functional arm could begin to be built. Any putative structure would have to be tested empirically, of course, to verify specificity. In addition, these antisense RNAs must be included in an overall structure that is carefully designed to maintain favorable reaction energetics such that the antiswitch is 'OFF' in the absence of succinate. Additional consideration must be paid to the antiswitch's sensitivity to alterations in pH, temperature, etc... These factors could all affect the delicate thermodynamics required for effective treatment. It is feasable to develop a fairly robust antiswitch that could functionally repress HIF1α in paraganglioma tumor cells.
B. Development of the aptamer:
There are no commercially available succinate-sensitive aptamers. The structure of succinate will have to be carefully considered and work would have to begin with building the aptamer from scratch.
C. Delivery Vehicle:
Administration of genetic material of any kind is difficult. Liposomes could be used to transfect the RNA antiswitch into paraganglioma tumor cells. The tumors are highly vascularized so global administration via injection would likely be most effective. Non paraganglioma cell-types are unlikely to be affected as they will not have abnormal accumulations in their nuclei. Further specificity could be conferred, however, by coating liposomes with antigens specific to somatostatin receptors, which have been shown to be endocytosis-mediating receptors which are fairly uniquely expressed by paraganglioma tumor cells during tumorigenesis (Herder, 2004).
REFERENCES
Amar L, et al: Succinate dehydrogenase B gene mutations predict survival in patients with malignant pheochromocytomas or paragangliomas. Journal of Clinical Endocrinology and Metabolism 2007, 92(10):3822-3828.
Asikainen TM, et al: Activation of hypoxia-inducible factors in hyperoxia through prolyl 4-hydroxylase blockade in cells and explants of primate lung. Proceedings of the National Academy of Sciences USA 2005, 102(19):10212-7.
Baris O et al: Transcriptional profiling reveals coordinated up-regulation of oxidative metabolism genes in thyroid oncocytic tumors. The Journal of Clinical Endocrinology & Metabolism 2004, 89(2):994-1005.
Bartrons R, and Caro J: Hypoxia, glucose metabolism and the Warburg’s effect. Journal of Bioenergetics and Biomembranes 2007, 39(3):223-229.
Baysal, et al: Sequence variation in human succinate dehydrogenase genes: evidence for long-term balancing selection on SDHA. BMC Biology 2007, 5(12):1741-1747.
Boedeker CC, et al: Malignant head and neck paragangliomas in SDHB mutation carriers. Otolaryngology-Head and Neck Surgery
Briere JJ, et al: Tricarboxylic acid cycle dysfunction as a cause of human diseases and tumor formation. American Journal of Physiology-cell physiology 2006, 291(6):C1114-C1120.
Cairns RA, et al: Metabolic targeting of hypoxia and HIF1 in solid tumors can enhance cytotoxic chemotherapy. Proceedings of the National Academy of Sciences of the United States of America 2007, 104(22):9445-9450.
Cheng VWT, et al: The iron-sulfur clusters in Escherichia coli succinate dehydrogenase direct electron flow. Journal of Biological Chemistry 2004, 281(37): 27662-27668.
Chrisoulidou A, et al: The diagnosis and management of malignant phaeochromocytoma and paraganglioma. Endocrine-Related Cancer 2007, 14(3):569-585.
Doring F, et al: Functional connections and pathways of coenzyme Q(10)-inducible genes: an In-silico study. IUBMB LIFE 2007, 59(10):628-633.
Ema, M, et al: Molecular mechanisms of transcription activation by HLF and HIF1 alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. Embrology Journal 1999, 18(7):1905-1914.
Frede S, et al: Regulation of hypoxia-inducible factors during inflammation. Methods in Enzymology 2007, 435:403-19.
Gaboli M, et al: Mzf1 controls cell proliferation and tumorigenesis. Genes and Development 2001, 15(13):1625-30.
Herder WW, Hofland LJ: Somatosensin receptors in Pheochromocytoma. Hormonal Research, 31:145-154.
Hoogewijs D, et al: From critters to cancers: bridging comparative and clinical research on oxygen sensing, HIF signaling, and adaptations towards hypoxia. Integrative and Comparative Biology 2007, 47(4):552-577.
Hsieh, YH, et al: Suppression of tumorigenicity of human hepatocellular carcinoma cells by antisense oligonucleotide MZF-1. Chinese Journal of Physiology 2007, 50(1):9-15.
Manelli M, et al: Genetics and biology of pheochromocytoma. Experimental and Clinical Endocrinology & Diabetes 2007, 115(3):160-165.
Morris JF, Hromas R, and Rauscher FJ: Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core. Molecular Cellular Biology 1994, 14(3):1786-95.
Oyedotun KS, and Lemire, BD: The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase – Homology modeling, cofactor docking, and molecular dynamics simulation studies. Journal of Biological Chemistry 2004, 279(10):9424-9431.
Pawlu C, Bausch B, Neumann HP: Mutations of the SDHB and SDHD genes. Familial Cancer 2005, 4(1):49-54.
Pollard PJ, et al: Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutation. Human Molecular Genetics 2005, 14(15):2231-9.
Qutub AA, et al: Three autocrine feedback loops determine HIF1 alpha expression in chronic hypoxia. Biochemica et biophysica acta-molecular cell research 2007, 1773(10):1511-1525.
Simon JM: Hypoxia and angiogenesis. Bulletin of Cancer Research 2006, 94:S160-5.
Szeto SSW, et al: Ubiquinone-binding site mutations in the Saccharomyces cerevasiae succinate dehydrogenase generate superoxide and lead to the accumulation of succinate. Journal of Biological Chemistry 2007, 282(37):27518-27526.
Woodward ER, and Maher ER: Von Hippel-Lindau disease and endocrine tumour susceptibility. Endocrine Related Cancers 2006, 13(2):415-425.