Abstract:
Systemic Lupus Erythematosus (SLE) is an autoimmune disease that can result in damage to many different tissues in humans ranging from joints to kidneys to cardiac muscle. The genetic causes of the autoimmune response characteristics of (SLE) are still incompletely understood. As part of the Davidson College Fall 2007 Biology 309 course investigation, one of the genes associated with SLE, known as Human La (an RNA binding protein involved in tRNA maturation), was analyzed with a variety of publicly available research data as well as public genomics and proteomics tools. Investigation of motifs indicated that the Human La gene contained a RUNX-1 binding site in the promoter region (JASPAR). Research into defects in RUNX-1 binding sites revealed that such defects in humans are associated with several autoimmune diseases, including SLE (Prokunina et al. 2002). Human La has an ortholog in yeast called LHP1 with similar functions (Ensembl Genome Browser, SGD). Analysis of yeast gene expression microarrays was conducted to uncover which genes have expression connected to LHP1. The analysis indicated that LHP1 has similar expression patterns to other genes associated with ribosomal function, but no obvious connections to SLE. Creation of a disease model indicated several areas for future research and suggests possible targets for treatment of SLE.
____________________________________________________________________________________________
Introduction to SLE and Human La:
Systemic lupus erythematosus, or SLE, is an autoimmune disease characterized by immune system attacks on healthy body tissues including the heart, skin, lungs, skeletal joints, kidneys, blood vessels and the brain. The disease is often hard to diagnose due to its varying symptoms and because it tends to flare up (especially in periods of stress) and then go away. The disease affects women more often than men and can run in families, indicating some sort of genetic predisposition. Drug treatments for SLE can run from simple anti-inflammatory drugs like ibuprofen and naproxen to corticosteroids to immunosuppressants in severe cases. For life threatening cases, a new treatment in clinical trials requires the extraction of bone marrow stem cells from the patient, the destruction of the immune system and then re-introduction of the stem cells into the bone marrow to “restart” the immune system (Mayo Clinic).
http://www.mayoclinic.com/health/lupus/DS00115/DSECTION=2
The protein known as human lupus antigen (human La) was originally named such because it was a target of the immune system in many cases of SLE (Ensembl Genome Browser). Many patients that suffer from SLE have anti-human La antibodies, which cause the immune responses to healthy tissues and the associated damage to those tissues. Human La is an important protein present in most cells and is involved in processes including tRNA maturation and tRNA chaperone activity (Pannone et al. 1998, Yoo et al. 1997). In this study, I investigated the connection between human La and SLE, searching specifically for a genetic cause of the prevalence of human La as an immune system target in SLE.
References:
ENSEMBL Genome Browser
http://www.ensembl.org/index.html
JASPAR
http://jaspar.cgb.ki.se/
Mayo Clinic Lupus Website
http://www.mayoclinic.com/health/lupus/DS00115/DSECTION=2
Prokunina, L. et al. 2002. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nature Genetics, Published online: October 28th 2002.
SGD (Saccharomyces Genome Database)
http://www.yeastgenome.org
Genomic Analysis of Human La:
This section focuses on the genomic location of Human La, the domains and motifs of the gene, the characteristics of the functional protein, similar proteins in other species and single nucleotide polymorphisms that could influence Human La function. This analysis was performed by using free-online tools and databases.
Genomic Location, domains and motifs:
The Human La gene is located on Chromosome 2 (Ensembl Genome Browser), contains three major conserved domains (NCBI), and several upstream motifs. Three major conserved domains were identified with NCBI’s Conserved Domain Search are the Lupus Antigen domain (of unknown function) and two RNA recognition motifs (Figure 1). These motifs are associated with the function of Human La, which is pre-tRNA binding and maturation (Ensembl Genome Browser).
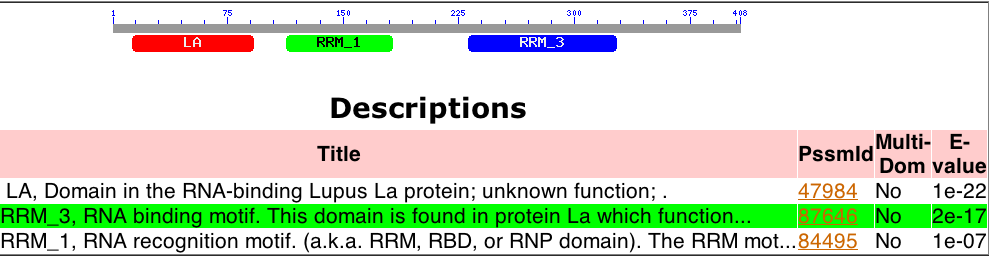
Figure 1. Conserved Domains of Human La. The gene has three main conserved domains: two of which are RNA recognition motifs (RRM1 and RRM3) and one of which is the Lupus Antigen domain, associated specifically with Human La.
The motif search yielded three major motifs in the 1,000 basepair section upstream from the coding portion of the Human La gene. The first of these motifs identified was the TATA Box, with a score of 7.710 (JASPAR). This motif is necessary for RNA polymerase binding and transcription of the gene. Another interesting motif was the Ar Sequence which is bound by an androgen receptor and which scored a value of 10.1. However, the most important motif identified was the RUNX1 binding motif, with a score of 9.296, which is bound by the transcription repressor RUNX1 (JASPAR). This protein has been implicated in many forms of autoimmune disease, like SLE, and therefore a binding domain for RUNX1 was of immediate interest (Helms et al. 2003, Prokunina et al. 2002, Tokuhiro et al. 2003).
Protein Structure and Function of Human La:
The protein structure and function was analyzed with several tools. A Kyte-Doolittle hydrophobicity plot with a window size of 20 found no membrane spanning portions of Human La, while the on the Hopp-Woods Scale, the protein was found to have several hydrophilic sections (Kyte-Doolittle Hydrophobicity Plots). Thus, it was determined that the protein was not a transmembrane protein. Additional analysis with PREDATOR found no major patterns in the protein with the exception of a large alpha-helix between the RRM1 and RRM3 conserved domains. Further research indicated that Human La is a very important protein for the cell process of tRNA maturation (Enembl Genome Browser).
Single Nucleotide Polymorphisms, or SNPs, of Human La were investigated. 27 variations were identified, of which only four caused non-synonymous coding (Ensembl Genome Browser). Of those four variations, one is a frameshift mutation and another is an essential splicing site mutation. These mutations were identified as being capable of eliminating Human La function. Unfortunately, since SLE is caused by Human La being an immune system antigen and not by “loss of function”, these SNPs were not of much relevance to the investigation. Additionally, no SNPs were present in any of the upstream motifs identified by JASPAR, including the RUNX1 binding site.
Orthologs of Human La:
Orthologs of Human La were identified in several species through Ensembl Genome Browser and then were analyzed using Clustal W. The resulting phylogenetic tree of the relatedness of the gene in different species matched the phylogenetic relationships of the species (Figure 2).

Figure 2. Cladogram based on the genetic similarity between orthologs of Human La. Note that it follows predicted evolutionary relationships (with the exception of the pufferfish).
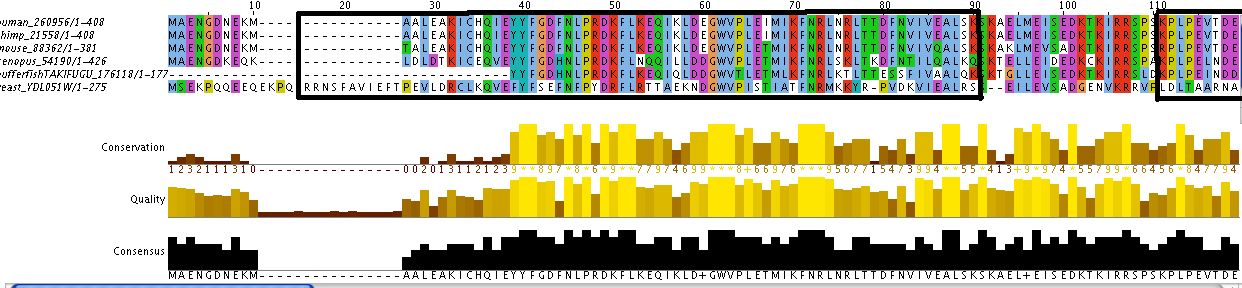
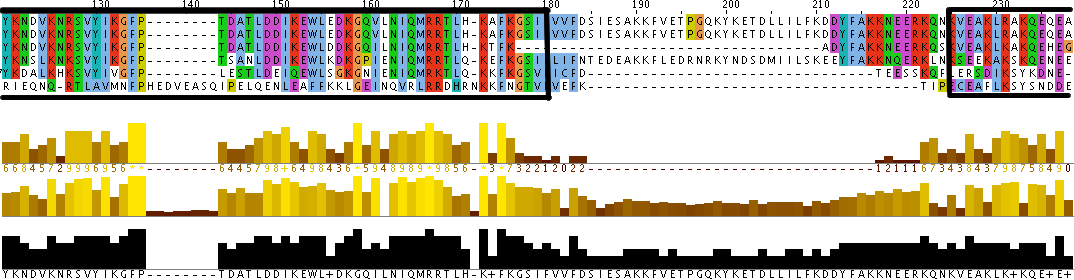
Additionally, the individual protein codes were matched up against each other and the conserved domains were mapped (Figure 3). This analysis indicates fairly good conservation between most species in the Lupus Antigen domain and moderate conservation among the others. This indicates that other species, like S. cerevisiae, are good model species for studying Human La. However, S. cerevisiae does not have any orthologs to the protein RUNX1, despite having RUNX1 putative binding domains. Thus, while a good model, the ortholog for Human La in yeast is not a perfect copy of the human protein.
 |
 |
 |
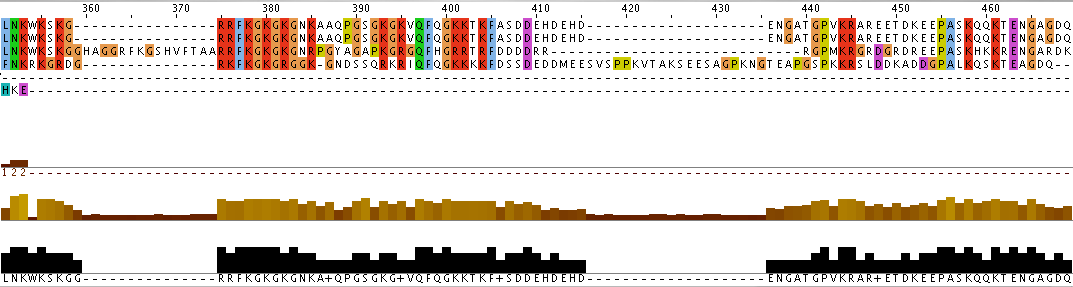 |
Figure 3. Jalview of Human La and other species orthologs with superimposed conserved domains. Note that conserved domains coincide with higher areas of conservation between species. Click to enlarge.
References:
ENSEMBL Genome Browser
http://www.ensembl.org/index.html
EBI Tools: Clustal W
http://www.ebi.ac.uk/Tools/clustalw/
Helms, C. et al., 2003. A putative RUNX1 binding site variant between SLC9A3R1 and
NAT9 is associated with susceptibility to psoriasis. Nature Genetics. Published online: November 9th 2003.
JASPAR
http://jaspar.cgb.ki.se/
Kyte-Doolittle Hydrophobicity Plots
http://www.vivo.colostate.edu/molkit/hydropathy/index.html
NCBI Conserved Domains Search
http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
PREDATOR: Secondary Structure
http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html
Prokunina, L. et al. 2002. A regulatory polymorphism in PDCD1 is associated with
susceptibility to systemic lupus erythematosus in humans. Nature Genetics, Published online: October 28th 2002.
Tokuhiro, S. et al., 2003. An intronic SNP in a RUNX1 binding site of SLC22A4,
encoding an organic cation transporter, is associated with rheumatoid arthritis. Nature Genetics: Published online: November 9th 2002
S. Cerevisiae Expression of Human La Ortholog:
The S. cerevisiae LHP1 gene is very similar in its conserved domains to Human La. Additionally, LHP1 has a similar function in yeast: that of tRNA maturation and chaperoning (Lin-Marq et al. 1995, Pannone et al. 1998, Yoo et al. 1997, Yoo et al. 1994). Therefore, studying the gene expression of LHP1 can be helpful in understanding expression patterns of Human La. During this part of the investigation, microarray data from the Hoopes laboratory at Washington University in St. Louis was used to investigate which genes have similar expression profiles to LHP1. Additionally, data was collected from the yeast genome database (SGD) and the TRIPLES database.
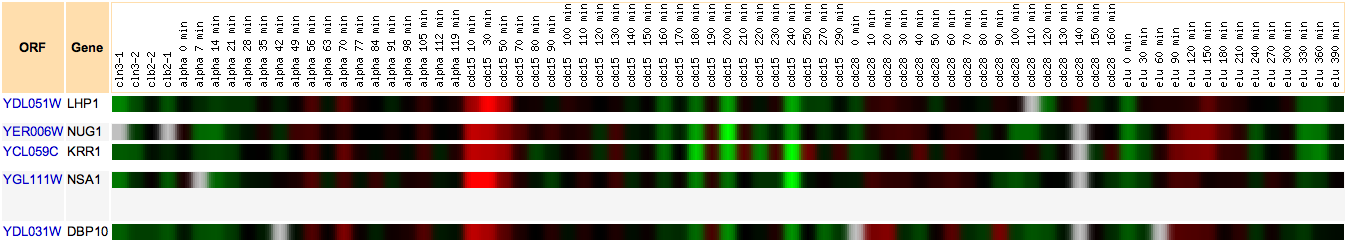
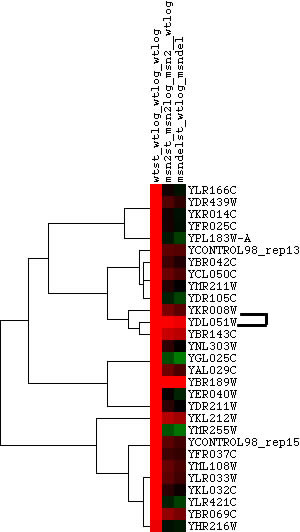
Hoopes laboratory data from strains of msn2/msn4 deletion mutants and wild-type yeast grown in log phase and stationary phase were used for the initial analysis. Msn2 and msn4 are transcriptional activators that function under stress conditions. Since stress in humans can cause outbreaks of SLE, it was hypothesized that there might be some connection between LHP1 and the stress response in yeast. Since LHP1 is a gene associated with tRNA and therefore translation, it was hypothesized that its expression should be elevated in periods of increased translational action. The data was analyzed using Magic Tool and then the proteins that had similar expression profiles to LHP1 were identified and are shown in Figure 1. As can be seen from Figure 1, the majority of proteins that clustered with LHP1 are connected to translational activity.
 |
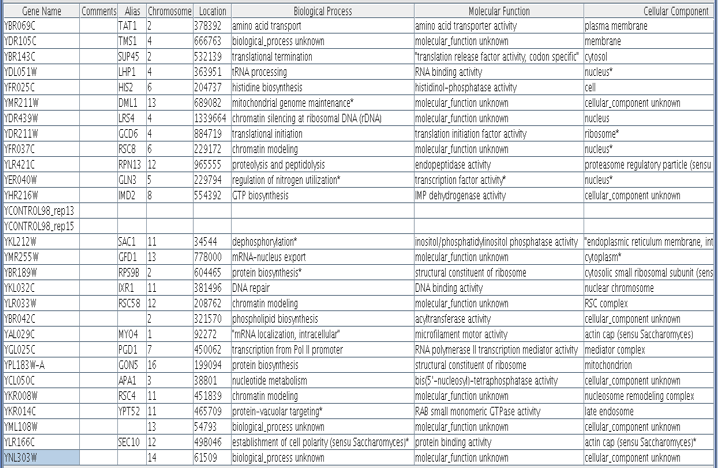 |
Figure 1. Proteins with similar expression ratios to LHP1 / YDL051W, the Human La ortholog, in msn2/msn4 deletion and wild-type yeast. YDL051W indicated by black marks on figure. The gene ontologies of the genes that cluster with YDL051W, displayed in table, vary but tend to be involved in ribosomal function and translation or transcription. This is not surprising, since LHP1 is involved in tRNA processing.
Next, data was collected from the Yeast Genome Database so that other growth conditions could be analyzed. Growth conditions of log-phase growth versus sporulating yeast and cell cycle expression versus a baseline gene expression were analyzed. Under conditions of sporulation and through different parts of the cell cycle, translational machinery is needed to different degrees. It was hoped that this would help identify more proteins that had similar expression to that of LHP1. As can be observed in Figure 2 and Figure 3, the only genes that consistently cluster with LHP1 are those involved with some part of translation or, oftentimes, transcription.

Figure 2. Proteins with similar expression ratios to LHP1 / YDL051W, the Human La ortholog, during Sporulation and log-phase growth. In this study there are no protein groups that seem to cluster with LHP1 with the exception of RNA processing proteins and proteins associated with translation.
Figure 3. Proteins with similar expression ratios to LHP1 / YDL051W, the Human La ortholog, during various stages of the cell cycle. Genes that cluster with LHP1 throughout the cell cycle are genes in the functions of rRNA and tRNA processing and ribosome biogenesis. Greatest induction occurrs at the beginning of the cell cycle for the cdc15 strain, when tRNA processing should be high due to increased translation. Click to enlarge.
Finally, yeast macroarray data was analyzed from the TRIPLES website to identify if the previous observations about sporulation indicated a necessity for LHP1 in the process of sporulation. The TRIPLES data indicates that yeast express LHP1 more during periods of vegetative growth rather than during sporulation. However, this could be simply due to the narrow period during which LHP1 is highly expressed during sporulation.
The main contribution of LHP1 to understanding Systemic Lupus Erythematosus in humans may be that LHP1 is sometimes expressed in periods of stress. SLE often flares up as well during periods of stress. There might be some connection between over-expression of Human La and increased autoimmune response.
References:
Lin-Marq N and Clarkson SG 1995. A yeast RNA binding protein that resembles the human autoantigen La. J Mol Biol 245(2):81-5.
Magic Tool
http://bio.davidson.edu/projects/magic/magic.html
Pannone BK, et al. 1998. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J 17(24):7442-53.
Saccharomyces Genome Database
www.yeastgenome.org
TRIPLES
http://ygac.med.yale.edu/triples/default.htm
Yoo CJ and Wolin SL 1997. The yeast La protein is required for the 3' endonucleolytic cleavage that matures tRNA precursors. Cell 89(3):393-402.
Yoo CJ and Wolin SL 1994. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol 14(8):5412-24.
Disease Model For Systemic Lupus Erythematosus:
Wild-Type Model:
The model of the non-SLE expression of Human La can be seen in Figure 1. This model was constructed through information gathered through the primary literature (see References) and the Human Interactome Map. Most of the parts of the pathway illustrated in the model are supported from the literature. Three important assumptions have been made. The first is that RUNX-1 is actually binding its binding site on the upstream end of Human La and is regulating the gene. While this deduction is fairly sound, especially since there seems to be a connection between failure of RUNX-1 regulation and autoimmune disease, there is no literature per-se that supports it. Additionally, the Human Interactome Map suggests that Signal Recognition Particle 54 may transport Human La. This might be one pathway through which Human La is exported out of the cell and thus targeted by the immune system. There is also a missing protein (labeled “?”) that must be present to transport Human La into the nucleus.
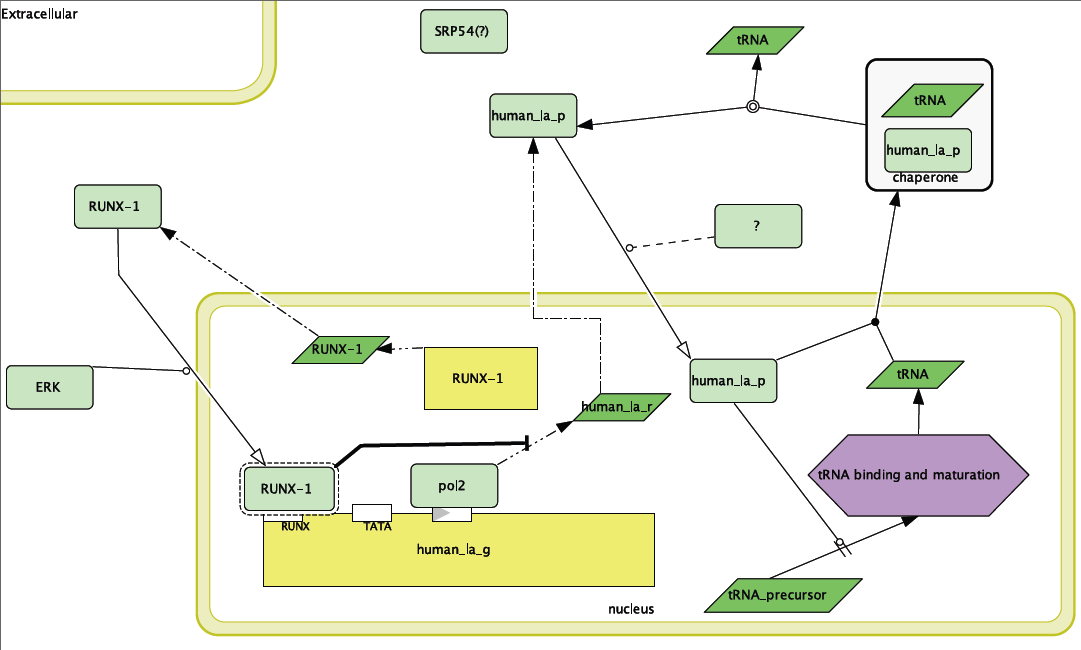
Figure 1. Wild-type model for Human La function. Note inhibition of transcription of Human La RNA by RUNX-1 binding.
Disease Model:
In modeling SLE, there were several difficulties. The first and foremost was that the disease is not associated with a mutation in the “disease” gene, but rather in the body’s response to that gene. Thus, previously uncovered mutations in Human La were not included in the model because, while they would be very detrimental to the health of the organism, they are not associated with the disease phenotype of SLE. In other words, the problem is that the immune system is responding to the proper protein in an improper fashion. Therefore, I focused the disease model on the question of how Human La is getting into a place where the immune system can target it. The disease model is presented in Figure 2. This model depends on some improper function of RUNX-1, whether that is improper transcription, phosphorylation or general function of the protein. Without RUNX-1 repression of Human La, the protein is over-produced and could either “leak out” or be transported out by SRP-54.
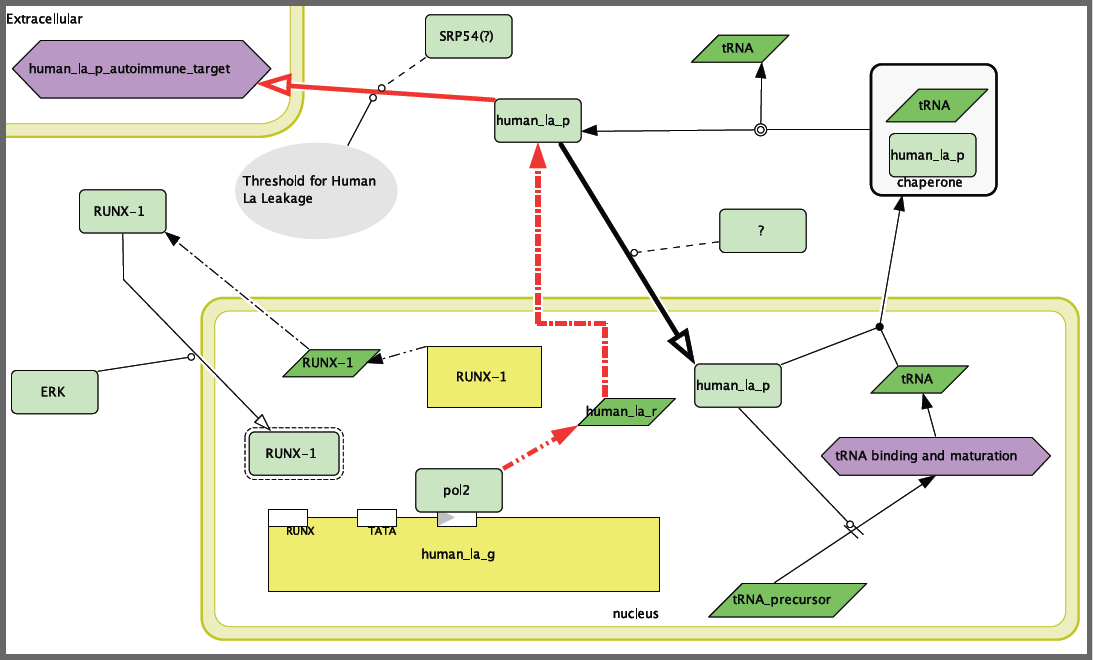
Figure 2. Systemic Lupus Erythematosus disease model. Note that RUNX-1 is not blocking transcription of Human La, leading to an overabundance of the protein and leakage into the extracellular space where Human La can be attacked by the immune system.
Future research:
The disease model contains several “holes” that must be addressed before the model can be accepted. The first and foremost is whether or not RUNX-1 is actually regulating Human La. This could be tested by using anti-sense RNA in human cell lines and then verifying if there is in fact over-expression and “leakage” of Human La in the absence of RUNX-1. Similarly, the importance of SRP54 in the pathway could be tested with the above experimental technique, by combining repression of both RUNX-1 and SRP54 and verifying if the “leakage” of Human La stopped. Additionally, the protein that transports Human La into the nucleus is unknown. I would take advantage of the homology in function of Human La and LHP1, the yeast ortholog, to use knockout yeast to identify what protein(s) are necessary for LHP1 localization to the nucleus. Most likely, orthologous proteins would be involved in the human process.
New treatments:
The very nature of SLE makes it a difficult disease to treat, especially with any kind of gene therapy. Since SLE is never localized to any particular area, it is difficult to target that specific area without influencing the entire patient. That said, in the many cases of SLE that mainly influence the skin, it should be possible to design a topical treatment, perhaps with liposomes containing RUNX-1, that could introduce RUNX-1 into RUNX-1 deficient skin cells to reduce transcription and therefore production of the aggravating antigen, Human La. This same treatment method might also be applicable to other proteins that as of now seem less promising, like SRP54.
References:
Alarcon-Riquelme, ME. 2004. Role of RUNX in autoimmune diseases linking rheumatoid arthritis, psoriasis, and lupus. Arthritis Res Ther. 6:169-173.
Bae SC, Lee YH. 2006. Phosphorylation, acetylation, and ubiquitination: the molecular basis of RUNX regulation. Gene. 336(1):58-66.
Bayfield MA, et al. 2007. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Molecular and Cellular Biology. 27(9):3303-3312.
Cell Designer
http://www.celldesigner.org/
ENSEMBL Genome Browser
http://www.ensembl.org/index.html
Harder, Ben. 2004. All roads lead to RUNX: several autoimmune diseases share one bad actor. Science News. April 3, 2004 Vol 165.
Human Interactome Map.
http://www.himap.org/
Mayo Clinic Lupus Website
http://www.mayoclinic.com/health/lupus/DS00115/DSECTION=2
Prokunina, L. et al., A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nature Genetics, Published online: October 28, 2002.