The Discovery Questions (DQ's) are not busy work or grade inflators. The DQ assignments are designed to help you
1. Digest the challenging material presented in this course.
2.
Keep your studying on track.
3. Prepare for in-class discussions and lab.
4. Train your brain for the exams, and ultimately train your brain to critically think about the complex cutting-edge integrative research that is Genomics, Proteomics and Systems Biology.
You are required to read the assigned pages and complete the assigned exercises before class. The writing format is casual (compared to exams answers). Please be as concise as reasonably possible. Do not sweat over composition, but if you take data from somewhere, do list the reference (no in-text citation is required). Answer the questions the best you can. Feel free to write wild speculations, point out database or link errors, places where you got lost in the database, etc. You will get feedback from me fairly quickly; definitely before the upcoming exam.
Important Instructions:
1. Answer Style: Here are some helpful tips on answering DQ's
Hypothesizing - when asked to formulate a hypothesis, frame it as an idea that can be tested. A speculation is "the gene might encode a product that has something to do with cell growth"; a testable hypothesis is "I hypothesize that expression of the gene is required for cell growth" (a test would be to delete the gene and look for changes in cell growth).
Reporting Database Searches - explain to me how you found your results. Comment on the logic you used in each key step. Be thorough, yet concise.
Screen Shots - screen shots (captured images from your computer screen) are helpful in showing search engine output, molecular images, etc. See the section on Screen Shots for details.
Be Thorough, Yet Concise - Brain dumping is the practice of unloading every bit of superficially associated bit of information you can think of onto paper. Competitive brain dumping is inflating the length of your answer to make it more impressive looking/ sounding than your classmates' answers. In your graded homework, brain dumping will receive a polite warning (i.e. "What you have up to this point is sufficient") but in your exams, brain dumping will be penalized (negative points). If you need tips on being thorough, yet concise, ask me for help.
2. Formatting: As a header, type your name, the title of the assignment (i.e. "Homework 1-1") and the date. This helps me stay organized, which prevents me from messing up your grade.
3. Deadline: Submit typed answers (i.e. Microsoft Word) to the assigned exercises to me via e-mail before 11 am, the day of class. I will not accept late work. You are allowed to drop two assignments per Unit. Use these allowances responsibly.
Abstract 1 should summarize the work completed during Unit 1.
Abstract 2 should include a brief recap of Unit 1 in the background and summarize the work completed during Unit 2.
The final writing assignment is your Final Report, which will cover work completed during all three units. It will include an abstract; therefore no Unit 3 abstract is assigned seperately.
Please do not exceed 250 words (not including title, name and affiliation). Include a title, your name, affiliation (as shown in the examples below) background/ significance of the work, procedure, results, conclusion, and discussion. Below is a very short made-up abstract to help you get started. A horrible version is on the left and a greatly improved version is on the right (my notes in brackets).
The First Lab Assignment Where We Looked for Genes in Shrew [technical name?] DNA [title is too open-ended and casual] Karmella A. Haynes Davidson College, Department of Biology, Davidson, NC 28036 Bioinformatics is really important for diagnosing and curing diseases. Lots of different species have had their genomes sequenced and these sequences are available online. The professor told us to find genes in a DNA sequence contig from the shrew. [significance is too broad and unconvincing] I got contig EEX000354647and we looked at it with the Ensembl Genome Broswer. We ran GenScan and looked at the bases and amino acids in the sequence. [We? There's just one author. The purpose of your work isn't very clear] The Genome Browser's data turned out to not be very good. GenScan and manual sequence gazing is better than trusting Ensembl. [please clarify]
|
Analysis of a Contig from Sorex araneus Reveals Critical Annotation Errors [Title is nicely worded as a general conclusion] Karmella A. Haynes Davidson College, Department of Biology, Davidson, NC 28036 The newly released sequence of Sorex araneus (the common shrew) offers opportunities for studies of DNA conservation across mammalian species. As of 2007, the current draft has only 2x coverage, thus there are many gaps between contigs and annotations are questionable. [Background is concise, significance is clear] As part of the Davidson College Fall 2007 BIO309 course investigation, I compared the current Ensemble Genome Browser annotations for contig EEX000354647 to my own GenScan analysis and manual intron-exon boundary annotation. [Procedure is clear] My analysis revealed the nature of some descrepancies in the official annotation. [general Results summarized] One of these discrepancies, due to a gap between EEX000354647 and the adjacent contig, omits a highly conserved exon which encodes a protein critical for proper calcium ion channel formation [key Result highlighted and described clearly]. These findings demonstrate that the annotations in the current S. araneus genome sequence draft should be considered with caution [Conclusion stated, helpful advice for the science community is a nice touch]. Further efforts to improve sequence coverage will allow proper annotation of this exon and subsequent gene conservation analyses. [Discussion is significant and helpful] |
Important Instructions:
You will submit an abstract of your lab work the week prior to giving your oral presentation. Due dates are highlighted yellow in the lab schedule (http://bio.davidson.edu/courses/genomics/lab.html). Send your abstract as an attachment to me via e-mail before the deadline.
Follow the instructions given on the cover page of your exam. You can e-mail me if you have technical questions concerning formatting, broken links, using Microsoft Word, etc.
In this lab, you will rigorously investigate a segment of the human genome using various sequence analysis techniques. You will then focus on a gene (within that segment) that is linked to a human disease. You will attempt to decipher the underlying mechanism of the disease using original data generated from microarray experiments, database exploration, cross-species sequence alignments, etc. Your research experience will culminate as a final report (web site).
You are required to carefully document your bioinformatics research. The purpose of your research notes is to log the progress of your investigation and to document the logic and techniques you used at each step so that you can compose a final report that describes your research in full at the end of the semester.
Important Instructions:
1. Overall organization: Keep printed pages in a 3-ring binder. Do not submit notes as electronic files. Notes can be typed, then printed. Feel free to include hand-written notations on notes after you've printed them.
2. Date: Be sure to include dates on each day's work
3. Outline Research Objectives: It's a good idea to begin each day's notes with an outline of the research objectives. These will remind you of the purpose of past exercises.
4. Background/ Lesson Information: Lab often includes a white-board lesson. You can keep these notes in a different section (or seperate notebook) or integrate them into your personal research notes.
5. Research Documentation: Your documentation (text notes) should answer these questions: How did I get my data? What is my data? What does it mean? During Unit 1, I will give explicit instructions on what you need to record in your notes (screen shots, questions to answer, etc.). Afterwards, you will be responsible for making decisions on effective research documentation.
6. Running List of References: It is critical that you keep a running list of references from the time you start your notebook until the end of the semester. Waiting until the last minute to compile a complete list of references for a semester's worth of work will drive you insane. Whenever you use published literature, cite the literature properly, not the URL where you found it. If your citation is a database or online tool, you can cite it as a Link/ URL. See "References and Citations" for details. If you are unclear about any citation/ reference, please ask me for clarification.
7. Submission: Submit your research notes (2% of your final grade) to me on the day of your oral Power Point presentation.
Oral Presentation 1 should describe the work completed during Unit 1.
Oral Presentation 2 should include a couple of slides to recap work from Unit 1 (as background) and describe the work completed during Unit 2.
Important Instructions:
1. Format: Microsoft Power Point. Include a title slide with a title, your name and affiliation (from the abstract). An acknowledgements slide is not required.
2. Time limit: 15 minutes, 5 minutes for questions (20 min. total). Be mindful of the time limit when creating slides (1 minute per slide is typical).
3. Be mindful of the audience: Good slides have more visual than textual information. Text should be large enough to see from across the room. Choose colors wisely (yellow text/ lines on a white background is not good).
| Grading Rubric: characteristics of an excellent talk | |
| Introduction (15 points) | The research question or problem is creative, well-defined and connected to prior knowledge in the chosen area of study. The research angle is creative. |
| Analysis Rationale and Procedure (15 points) | Not just a recapitulation of lab instructions, but a summary of major steps and key variations. Presentation has sufficient detail to understand variables (i.e. sequence input and filter parameters) and experimental design (where applicable). Rationale is appropriate and covers important (not trivial) considerations. |
| Results (15 points) | The data are analyzed and expressed in an accurate way. Statistical analysis to support or reject conclusions is present (i.e. E-values, threshold values, etc. where applicable). |
| Figures and Tables (15 points) | The figures and/or tables are appropriately chosen and well-organized; data trends are illuminated; key results are clearly marked. Figure labels are informative, clear and complete. |
| Discussion/ Conclusions (15 points) | Effectively ties together the results in a consistent and logical manner. Inferences an conclusions are insightful, novel and non-trivial. Next steps (future work) are logical and relevant. |
| Clarity (15 points) | The presentation is visually appealing and easy to follow. It draws in the audiene and keeps their attention. No distracting backgrounds or animations are included. |
| Fielding Questions (10 points) | Responses to the questions exhibit sound knowledge of the study and the underlying science concepts; the presenter exhibits poise, confidence and enthusiasm. |
FYI: The Speaking Center at Davidson College offers the services of trained student tutors to support speaking across the curriculum. Location: Chambers B39 in the north basement. Hours: Sunday through Thursday from 9-11 pm starting Sunday, September 9th
The final report (web pagee) will tie together the research completed during the semester. The style should be scientific and accurate, yet readable by the general public. To minimize time spent on design decisions (this can take forever) I will provide a template home page and .css file for you. You should focus more on content than bell and whistles. On the last day of lab, you will give a guided walk-through of your web site (projected on the big screen) to the class. Verbal Q&A will be included in your grade for this assignment.
Important Instructions:
Sections - Your web page will have 4 sections
1. Intro - this will include your abstract, and background
2. Gene - Unit 1 research
3. Expression - Unit 2 research
4. Disease Model - Unit 3 research
Note: Each section must end with a list of references.
Figures - Figures should summarize your research from each Unit. Create no more than 3 figures per Unit. You are expected to combine all the work into a comprehensive figure(s) in a logical and informative way. You can make use of good slides from your previous talks by taking screenshots in Power Point (with spell check deactivated). You can show large figures as thumbnails, and link the thumbnail to the full sized figure to show details. Don't make the figures so huge that the viewer needs to scroll around to see the whole thing.
Uploading - you may use any of the following:
1. Cyberduck (FTP)
2. Fetch (FTP)
3. Dreamweaver
Note: Please do not override the .css settings to change the header image or background color. Keep your content inside the "div" tab sections.
Editing the Template - view the template
Your page is provided as a template called "home.html" in your folder. I have provided a nifty javascript to magically transform the 4 sections of your page into tabs for easy navigation. All you have to do is fill in the sections with your content.
You may earn extra credit by attending seminars relevant to genomics and submitting a written summary.
Important Instructions:
1. Attend the seminar (you will receive dates and times via e-mail).
2. Ask the speaker at least one interesting question. Write down your question and the speaker’s answer.
3. Submit the following written assignment via e-mail within one week after the seminar: Your name and the date you completed the assignment; the seminar's title, speaker's name and date; your question(s), the speaker’s answer(s); a brief (~5 sentences) summary of the talk from a genomics perspective. Please submit this within a week following the talk.
Links to the databases (i.e., OMIM, NCBI, PDB, etc.) are listed at The Genomics Place (http://wps.aw.com/bc_campbell_genomics_2/). Click Web Links in the left hand menu to access a full list of links. If you find a broken link, report it to me via email. See the list of corrected links.
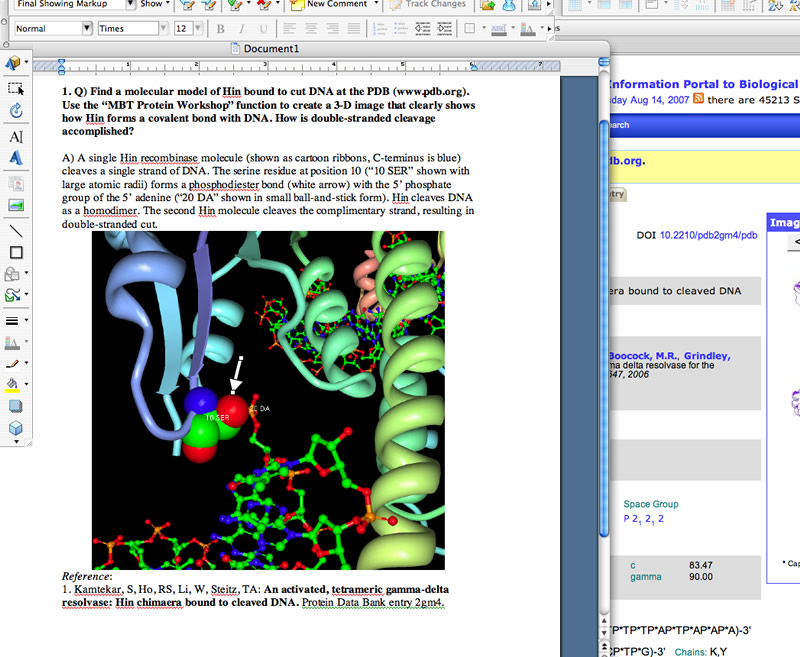
Not all Discovery Questions and exam questions require screen shots, BUT there are cases in which a screen shot is extremely effective. Real bioinformatics work involves looking at data that is often presented as molecular images, selected groups of output captured from huge databases, graphs generated on the fly, etc. that appear in your internet browser window. In genomics, an image is worth a thousand bits of data. Instead of decribing the output in words, use a program like "Grab" (a Mac utility) or "Snaps Pro" to capture the images. Save the image as a tiff or jpeg file, then insert the image into your document.
Shown below is an example of good use of a screen shot. The image is a snapshot of a student's computer screen where she is answering a question about enzyme structure and function. Notice how the question is presented first, the text answer concisely describes the image and states the answer, the image is clear and shown at a reasonable size, a key feature is pointed out with an arrow (generated using a Draw tool) and the reference for the data (in this case a 3-D image) is shown below the image.

Reference formatting varies in different courses, journals, etc. For the sake of consistency, for this class use the format shown in the examples below (from the Journal of Biological Engineering [http://www.jbioleng.org/info/instructions/]). References should be numbered and listed in alphabetical order.
Article within a journal |
1. Koonin EV, Altschul SF, Bork P: BRCA1 protein products: functional motifs. Nat Genet 1996, 13:266-267. |
Article within a journal supplement |
2. Orengo CA, Bray JE, Hubbard T, LoConte L, Sillitoe I: Analysis and assessment of ab initio three-dimensional prediction, secondary structure, and contacts prediction. Proteins 1999, Suppl 3:149-170. |
| In press article | 3. Kharitonov SA, Barnes PJ: Clinical aspects of exhaled nitric oxide. Eur Respir J, in press. |
| Published abstract | 4. Zvaifler NJ, Burger JA, Marinova-Mutafchieva L, Taylor P, Maini RN: Mesenchymal cells, stromal derived factor-1 and rheumatoid arthritis [abstract]. Arthritis Rheum 1999, 42:s250. |
| Article within conference proceedings | 5. Jones X: Zeolites and synthetic mechanisms. In Proceedings of the First National Conference on Porous Sieves: 27-30 June 1996; Baltimore. Edited by Smith Y. Stoneham: Butterworth-Heinemann; 1996:16-27. |
| Book chapter, or article within a book | 6. Schnepf E: From prey via endosymbiont to plastids: comparative studies in dinoflagellates. In Origins of Plastids. Volume 2. 2nd edition. Edited by Lewin RA. New York: Chapman and Hall; 1993:53-76. |
| Whole issue of journal | 7. Ponder B, Johnston S, Chodosh L (Eds): Innovative oncology. In Breast Cancer Res 1998, 10:1-72. |
| Whole conference proceedings | 8. Smith Y (Ed): Proceedings of the First National Conference on Porous Sieves: 27-30 June 1996; Baltimore. Stoneham: Butterworth-Heinemann; 1996. |
| Complete book | 9. Margulis L: Origin of Eukaryotic Cells. New Haven: Yale University Press; 1970. |
| Monograph or book in a series | 10. Hunninghake GW, Gadek JE: The alveolar macrophage. In Cultured Human Cells and Tissues. Edited by Harris TJR. New York: Academic Press; 1995:54-56. [Stoner G (Series Editor): Methods and Perspectives in Cell Biology, vol 1.] |
| Book with institutional author | 11. Advisory Committee on Genetic Modification: Annual Report. London; 1999. |
| PhD thesis | 12. Kohavi R: Wrappers for performance enhancement and oblivious decision graphs. PhD thesis. Stanford University, Computer Science Department; 1995. |
| Link / URL | 13. The Mouse Tumor Biology Database [http://tumor.informatics.jax.org/cancer_links.html] |
Cite references within the text by including the last name of the first author (if applicable) followed by the year of the publication in square brackets. For web links, include the title of the link in parenthesis. The example below demonstrates proper citation:
| Previous research has demonstrated that a repetitive element called hoppel, or 1360, contributes to pericentric silencing of an adjacent reporter gene [Haynes 2006]. The current Ensembl annotation of the D. melanogaster genome includes a track for repetitive elements, which shows copies of 1360 dispersed throughout chromosome four [Flybase]. |