Review of: The phenotypic legacy of
admixture between modern humans and Neandertals
Summary
This research conducted by Simonti et al sought to reveal the relative impact of interbreeding between
Homo Neanderthalensis and
ancestral Homo sapiens
approximately 50,000 years ago has on modern human physiology. To do this,
researchers analyzed data from ~28,000 adults of European ancestry,
combining genotyping data obtained from genome-wide arrays with phenotypic
data obtained from the Electronic Medical Records and Genomics (eMERGE)
network, an online consortium for electronic health records. These data
were referenced against a collection of ~135,000 Neanderthal specific
single-nucleotide polymorphisms (SNPs) identified by statistical
computation. By searching for correlations between “Neanderthal alleles”
identified in the genotyping data and phenotypic traits from the eMERGE
data, researchers sought to identify phenotypes associated with
Neanderthal DNA.
Conclusions
Analysis revealed functional associations between
Neanderthal alleles and several different phenotypes relating to
depression, metabolism, addiction, skin, and the immune system. The
researchers attributed these particular phenotypic associations to the
evolutionary past of Homo
Neanderthalensis, stating that the hominid might have developed such
phenotypes due to selection pressure as the species migrated to higher
latitudes. The researchers further support this conclusion by clarifying
that many of these associated phenotypes are directly influenced by
sunlight exposure, and that phenotypes that are disadvantageous today in
modern humans of European descent may have conferred a selective advantage
for ancestral populations as they migrated north.
Personal Assessment
In all, this study employed a novel technique that
uncovered new insights into the evolutionary mysteries of the human
genome. The researchers demonstrated the power and utility of the eMERGE
database, especially when complexed with genotypic data for each patient.
There are important limitations to this technique, though, most notably in
that only clinical phenotypes can be examined and mined given the limited
scope of the electronic health records. Nevertheless, this team
demonstrated a powerful method while simultaneously revealing novel
genotypic associations that were previously unknown.
Figures
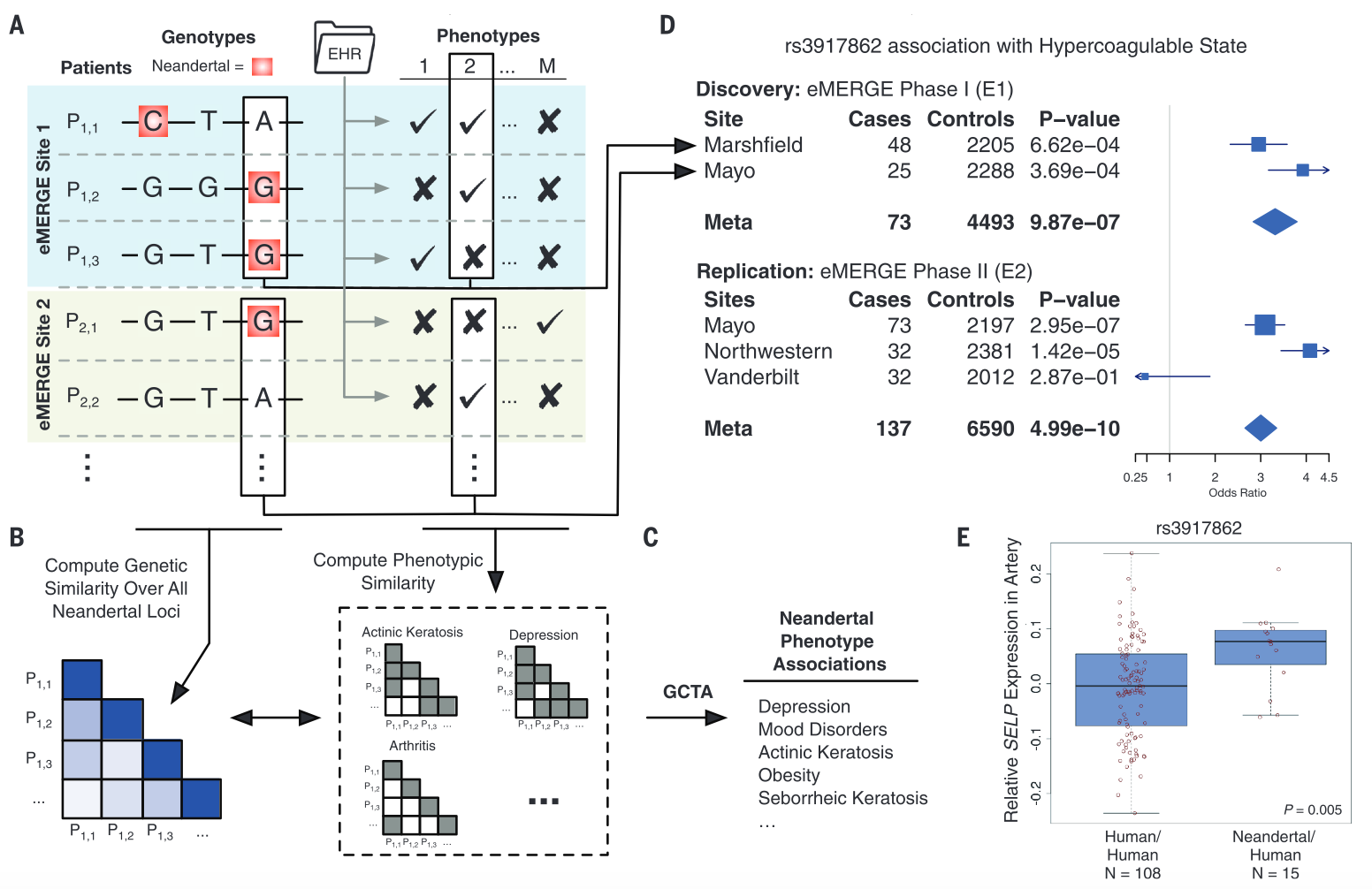
Figure 1
This schematic outlines the overall procedure that the researchers followed in this study. (A) First, results from the eMERGE database were compared to genotyping data for ~28,000 individuals of European descent, and SNPs attributed to Neanderthal ancestry were identified. (B-C) After compiling these results, Neanderthal phenotype associations were determined by comparing the data across all Neanderthal loci and searching for correlations. (D-E) Two different identified allelic associations are then demonstrated, providing evidence for the power and utility of the technique.

Table 1
This first table portrays eight different phenotypic associations identified by employing the research method shown in Figure 1A-C. The extent to which the Neanderthal alleles are responsible for each phenotype is calculated and listed under risk factor along with the associated p-value. The three different columns (E1, E2, and E2 two-GRM) refer to multiple analyses conducted on this dataset. The researchers specifically highlighted the first three phenotypes as particularly significant even when non-Neanderthal variants were controlled for.
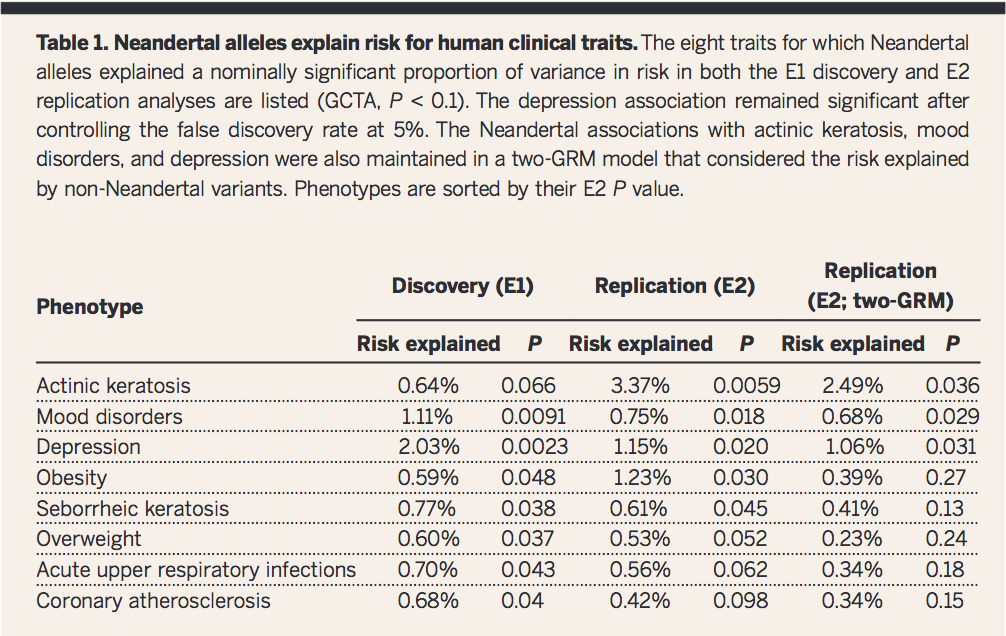
Table 2
This second table highlights four Neanderthal SNPs that correlated significantly with a particular phenotype observed in the eMERGE data. The flanking genes for each SNP are also reported, providing a putative candidate gene for each phenotype. Results and statistical results are provided for each of the individual analyses.
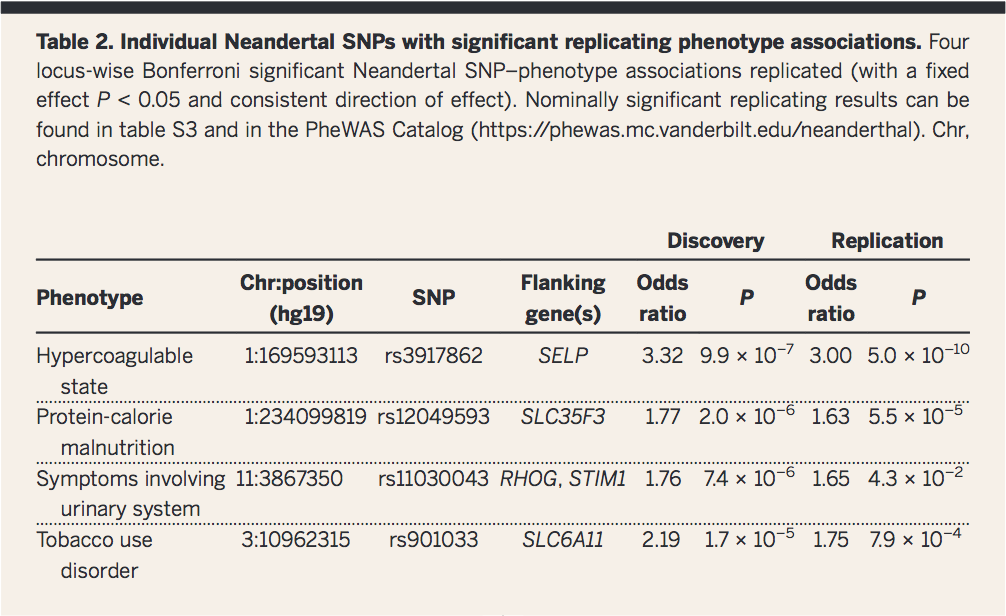
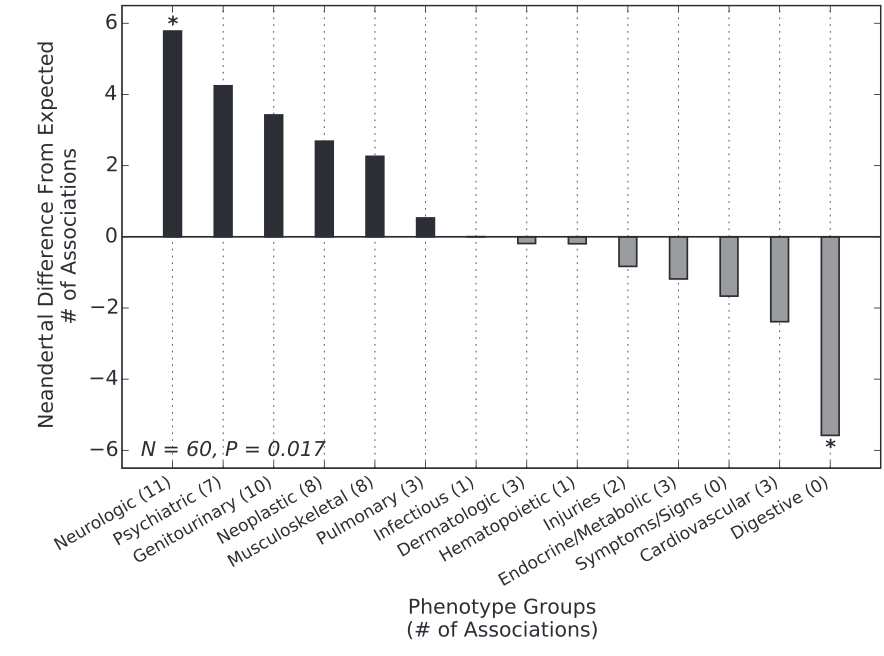
Figure 2
This graph classes the individual SNP associations in distinct phenotypic categories and compares the Neanderthal SNP categories with a set of non-Neanderthal sites that demonstrated similar allele frequency. A greater positive distance from the baseline of zero signifies a stronger association between Neanderthal SNPs and a certain phenotype. A greater negative distance signifies an inverse association. Significant associations with Neanderthal SNPs are observed with neurological and psychiatric phenotypes, and a significant disassociation is observed with digestive phenotypes.
Email Questions or Comments: jastewart@davidson.edu.
© Copyright 2016 Department of Biology, Davidson College, Davidson, NC 28035