GCAT
FAQ
1) What is GCAT
and what is its mission?
What are microarrays?
2) How do I join GCAT?
How much does it cost?
3) What species does GCAT
have?
How can I request microarrays for my undergraduates?
Can GCAT
get microarrays for my favorite species?
4) How do I pay for my DNA microarrays?
How do I store the microarrays?
5) Where does the money go?
6) What do I need to perform a
DNA microarray experiment?
How can I isolate yeast RNA?
How do I know if
my isolated RNA is any good?
7) How do I get my microarray scanned?
8) How do I collect my raw data?
How do I process the electronic data?
9) What additional information do
I need?
10) How do I analyze the results?
Can you show me a couple outcomes from microarray studies?
What are the capabilities of microarrays?
11) Why did my array have a gradient
of dye from one side to the other?
12) Why did my array have bright
background and black spots?
13) Why was my array pitch black?
14) What are all these big, out
of focus circles on my slide?
15) Why are the images grayscale
instead of color images?
16) How can I get my student's
results posted on the GCAT
web site?
17) Can GCAT
link to my course web site?
18) How can I contribute towards
GCAT
activities?
19) Are there any workshops to
get better at doing DNA microarrays?
20) Are there non-RNA methods I
could use instead?
21) Are there any raw data files
I can use for a dry lab exercise?
22) Are there good papers that
my students and I can use and also access the data?
23) Why wasn't this available when
I was a student?
24) This looks interesting but
I have no idea how to proceed. Where do I start?
1) What is GCAT
and what is its mission? What
are microarrays?
2) How do I join GCAT?
How much does it cost?
Joining GCAT
costs nothing if you do not want to use DNA microarrays from GCAT
or use a GCAT
scanner. To join, the best place to start is signing up for the listserv
called
GCAT-L.
This is how all GCAT
communication begins, including the time when requests for microarrays are
being accepted. You might want to attend a free GCAT workshop to learn how
to perform experiments and analyze data.
3) What species does
GCAT
have? Can GCAT
get microarrays for my favorite species?
GCAT
tries to obtain microarrays for these species:
-
Yeast (S. cerevisiae; Washinton
Univ., St. Louis)
- Yeast
bar code microarrays (No Longer Available)
-
E. coli (University
of Alberta)
-
Arabidopsis (Univ. AZ) Only 31 available, first come first served
- Chlamydomonas (Stanford Univ.) No Longer Available
-
Maize (Univ. AZ) No more requests, all chips pre-ordered
- soy beans (Univ. of Illinois) All requests due by April 29, 2011
- tomato (Cornell Univ.) Only 53 available, first come first served
- rice (Univ. AZ) No more requests, all chips pre-ordered
-
Mouse (Washinton Univ., St. Louis)
-
Rat (Washinton Univ., St. Louis)
-
-
Fly (D. melanogaster; Univ. Health Network, Toronto)
- C. elegans (Washinton Univ.,
St. Louis) No Longer Available
- Chicken (Univ. AZ)
- zebra fish (Agilent) No Longer Available
Due to GCAT's
recent HHMI support, we
will be able to produce our own yeast microarrays! All other species will
continue to be supplied by academic labs, though now GCAT
will be able to compensate the labs for their generosity. Since we obtain
most microarrays from labs that have their own research interests, we cannot
guarantee non-yeast species or number of microarrays. However, GCAT
always tries to meet the needs of its members.
Pricing Structure and Availability
A) Schools supporting GCAT
with their HHMI money will get first options and will not need to pay any
more money to obtain the microarrays.
B) All other schools will need to pay the standard
rate of $60 for the first microarray and $20 for each additional microarray,
per semester. $40 dollars covers FedEx shipping and packaging. The $20
was mandated by NSF and pays for the scanner's service contract. Malcolm
does not receive any compensation for this work, nor does Davidson College.
We can only accept checks.
C) I will do my best to match all requests, but
for non-yeast microarrays, there may be a limitation to the number of microarrays
any one school can receive through GCAT.
D) The species we have available this coming year are:
-
-
Yeast (S. cerevisiae; Washinton
Univ., St. Louis)
- Yeast
bar code microarrays (No Longer Available)
-
E. coli (University
of Alberta)
-
Arabidopsis (Univ. AZ) Only 31 available, first come first served
- Chlamydomonas (Stanford Univ.) No Longer Available
-
Maize (Univ. AZ) No more requests, all chips pre-ordered
- soy beans (Univ. of Illinois) All requests due by April 29, 2011
- tomato (Cornell Univ.) Only 53 available, first come first served
- rice (Univ. AZ) No more requests, all chips pre-ordered
-
Mouse (Washinton Univ., St. Louis)
- Rat (Washinton Univ., St. Louis)
-
-
Fly (D. melanogaster; Univ. Health Network, Toronto)
- C. elegans (Washinton Univ.,
St. Louis) No Longer Available
- Chicken (Univ. AZ)
- zebra fish (Agilent) No Longer Available
E) What do I need now is to hear from you? Tell me what
you want using these guidelines.
F) If you have a new species you'd like to work
with, then provide Malcolm Campbell
with the contact information of a lab you think might be willing to share
with GCAT.
Malcolm will contact them to see if this species can be added to the list.
G) If your institution requires a W-9 form, please
enter this information:
- Check the N - Nonprofit Organization box and
enter the TIN# 562161106
4) How do I pay for my
DNA microarrays? How do I store the microarrays?
GCAT
is run by Malcolm Campbell as a service to the undergraduate education community.
He has a non-profit status bank account set up in Davidson that handles
all the money. Therefore, the only way to accept money is in the
form of a check (personal or institutional). Sorry, GCAT
cannot accept Purchase Orders (PO's) or credit cards. Make checks payable
to:
GCAT
c/o Malcolm Campbell
Biology Dept.
Box 7118
Davidson College
Davidson, NC 28035
Store your microarrays at room temperature in
a dessicator.
5) Where does the money
go?
This is a good question. The money ($60 for
the first microarray per species + $20 for each additional microarray) pays
for the FedEx delivery of the microarray and the scanning. The GCAT
scanner was purchased on an NSF grant where GCAT
promised to charge $20 per scan to pay for the service contract (currently
$7,000 a year). Malcolm Campbell does not receive any compensation for his
services, nor does Davidson College. All money is used for GCAT
expenses.
6) What do I need to
perform a DNA microarray experiment?
You need the following equipment:
-
Standard molecular biology small equipment
-
Incubation chamber (water or air)
-
Compressed air or centrifuge that holds
50 mL conical tubes to dry the slides
-
Capacity to isolate and manipulate RNA
(many choices of methods)
- cDNA production and labeling kit (many choices but special
discounts on 3DNA)
-
Access to FedEx or UPS next morning
delivery (if you want GCAT to scan your chips)
-
Computer with at least 512 MB RAM
-
To run GeneSpring or Scanalyze, you need
a PC: to run MAGIC Tool, you can use
Linux, PC or Mac OSX
-
Access to FTP program and the internet.
-
Undergraduate students only - no graduate
students for wet lab or GeneSpring.
7) How do I get my microarray
scanned?
First, you must contact a GCAT
scanning center one week in advance to verify the time is acceptable. You
must inform the scan center how many microarrays you intend to send and
what type of microarrays you are sending. Keep in mind that each microarray
takes approximately 30 minutes to scan, so it is helpful to space out your
microarrays rather than sending more than 5 at a time.
When you pay $20 per microarray, you have paid for the scanning
service. If you obtain your own microarrays but want GCAT
to scan them, the charge is $20 per scan. GCAT
only scans microarrays for undergraduate research or course work.
You must overnight express by FedEx, UPS, DHL, etc. (not
US Mail) your microarrays to GCAT.
Do not send them by standard overnight delivery because they arrive too
late in the day to scan them on the same day.
Scan Centers:
| Another Choice |
Another Choice |
Another Choice |
Another Choice |
Emily Dixon
Assistant Professor
Departments of Biology and Chemistry
St. Lawrence University
23 Romoda Drive
Canton, NY 13617
(315) 229-5671 |
Mark
Gallo
Dept. of Biology
Niagara University
NY 14109
(716) 286-8247
|
Kam Dahlquist
Dept of Biology
Loyola Marymount Univ.
1 LMU Drive,
MS 8220
Los Angeles, CA 90045-2659
Tel: 310-338-7697
|
Richard Musser
Waggoner 372
1 University Circle
Dept of Biological Sciences
Western Illinois University
Macomb, IL 61455
(309) 298-1096
|
8) How do I collect my
raw data? How do I process the electronic
data?
Your scanned slides will appear on the ISB
FTP server. To access the files, you need to use an FTP program, not a
web browser. This means you CANNOT use Internet Explorer, or Netscape,
or Safari, or FireFox, etc. You must use a program such as Fetch, CuteFTP,
CyberDuck, etc.
The URL is ftp.systemsbiology.net. The
usrname=gcat (use
only lower case letters; see example below). The password will be
sent to you by email from Malcolm
Campbell after you request it. You will have two tiff files for
each scanned slide. To learn more about
the tiff file format, go to this web site.
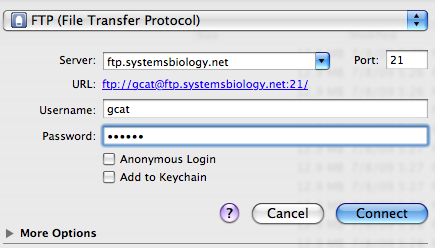
It is very important that you do NOT
give the username and password combination out to any students. GCAT
is a guest on the ISB server and we do not want to abuse this privilege.
9) What additional information
do I need?
You must be able to identify the gene located
at each spot (feature). For this, you need to know how the microarray was
printed (e.g. how many rows and columns per grid, how many different grids,
how many metagrids, how many times each gene was spotted and were gene spots
printed in adjacent pairs?). You will also need the genelist or godlist which
is a tab delimited text file (or converted to one via Excel) that lists each
gene on a separate line. This information must be supplied to you and you
can find most of this information at the GCAT
Protocols page. Orientation is also an issue which
you can learn about on this gridding page.
10) How do I analyze the
results? Can you show me a couple
outcomes from microarray studies? What
are the capabilities of microarrays?
GCAT
does not require you to use any particular method. Some faculty prefer Scanalyze
to quantify the spots and GeneSpring
to analyze multiple microarray data. Scanalyze is free but only works
on a PC. GeneSpring
has been free and also requires a PC. Each year, GCAT has to negotiate the
availability of GeneSpring.
You may want to use MAGIC
Tool which was written by GCAT
faculty and students and performs both halves of the analysis, though with
no bells and whistles. MAGIC Tool works
on Linux, PC and Mac OSX.
11) Why did my array
have a gradient of dye from one side to the other?
You did not mix the hybe solution well before
incubating. Read the Tips File for details.
12) Why did my array
have bright background and black spots?
This problem has a few causes. One is too much
SDS that did not get washed off. Other reasons include poor blocking or too
little heat while washing. Read the Tips
File for details.
13) Why was my array
pitch black?
The most common problem is poor quality RNA.
It is easy to degrade. The second most common problem is poor cDNA synthesis.
Finally, it helps to boil the microarray before hybridizing. This is required
if you are using cDNA (PCR product) spotted on the microarray. Read the Tips
File for details.
14) What are all these
big, out of focus circles on my slide?
The microarray scanner
at Davidson is not laser based. The out of focus blobs (typically circles)
are dust spects on the back side (non-DNA side) of your microarray. All microarrays
are blown clean with compressed gas, but some dust does not come off. Therefore,
it is especially important to use clean wash solutions, do not expose your
microarrays to lent-causing fabric such as kimwipes, and keep your microarrasy
in a dust-free box.
The scanner at Pomona is laser based and thus
does not produce the same spots. However,
each device has its tradeoffs, so neither one is perfect.
15) Why are the images
grayscale instead of color images?
All microarray scans are tiff files and all
tiff files are grayscale. The colors you see are generated by the software
once you tell the software which file is supposed to be red or green. Each
pixel on the tiff file can have a grayscale intensity of 1-128. This information
is used by the software to produce the numerical values and the color intensity.
16) How can I get my
student's results posted on the GCAT
web site?
If you want your students'
data posted too, just email Malcolm
Campbell with the URL and the description you want associated with the
link.
17) Can GCAT
link to my course web site?
Yes, email Malcolm
Campbell with the URL and the description you want associated with the
link.
18) How can I contribute
towards GCAT
activities?
You can help in many ways. First, participate
in the GCAT-L
listserv. When someone asks a question that you can answer, please reply to
the list (GCAT-L@davidson.edu)
with your answer. "It takes a community to hybe a chip."
Second, you can send URLs for course
sites and student data for posting on GCAT.
Third, you can share any presentations your
students make by emailing Malcolm
Campbell with the details and the description you want associated with
the information. GCAT
would be happy to mirror any PowerPoint files you care to share.
Fourth, you can send jpeg files for inclusion
in our photo gallary of GCAT
students and faculty. Your photo can be fun or more serious; your choice.
Fifth, you can volunteer to help coordinate a
species. By that, you might act as the go-to person for species-specific questions,
help communicate with the microarray supplier, etc.
Finally, you can cite GCAT
in any publications or lab manuals you produce. GCAT
is an all volunteer organization and we grow through your efforts at communicating
success and lesssons learned along the way.
19) Are there any workshops
to get better at doing DNA microarrays?
GCAT
has offered two hands-on workshops so far,
though no others are planned at this time. You should check with Cold
Spring Harbor's Dolan DNA Learning Center. There have been two MAGIC
Tool workshops as well.
20) Are there non-RNA
methods I could use instead?
Yes, there are two options. One is comparative
genome hybridizations where you label genomic DNA instead of cDNA. This allows
you to compare two similar species or strains, or mutant and wild-type isolates.
Do a PubMed
search to learn more. Here
is one example.
Malcolm Campbell has also developed some "teaching
chips" which use oligonucleotides as probes and cloned PCR products
as spotted targets. You could produce these "microarrays" by hand
if you wanted.
21) Are there any raw
data files I can use for a dry lab exercise?
Yes, GCAT
has several tiff files and expression files
you can use to get started on dry lab data analysis. GCAT
is also collaborating with Cybertory.org
as they continue to develop their synthetic tiff file production method.
22) Are there
good papers that my students and I can use and also access the data?
Yes, lots of them. A good starting place is
Stanford Microarray Database,
but you can find many others through PubMed. You might also check out the
Open Accesss journal PLoS.
23) Why wasn't GCAT
available when I was a student?
This is a multiple choice question:
24) GCAT
looks interesting, but I have no idea how to proceed. Where do I start?
First, sign up for GCAT-L
listserv. This will keep you informed on the news and what problems people
are working on. Once a year, a call goes out for microarray
requests. You must submit your request by the stated deadline in order
to be considered for microarrays in the coming academic year. Because GCAT
is an all volunteer organization, we cannot take requests year round.
Second, you can begin with dry-lab experiments.
You can use MAGIC Tool and analyze the practice
data or download real data sets online.
It turns out that the hard part is not collecting
data (though that's not easy), the hard part is data analysis. So you can
start there for free and see how it goes.
GCAT Home Page
Biology Home Page


© Copyright 2007 Department of Biology, Davidson
College, Davidson, NC 28035
Send comments, questions, and suggestions to: macampbell@davidson.edu
