
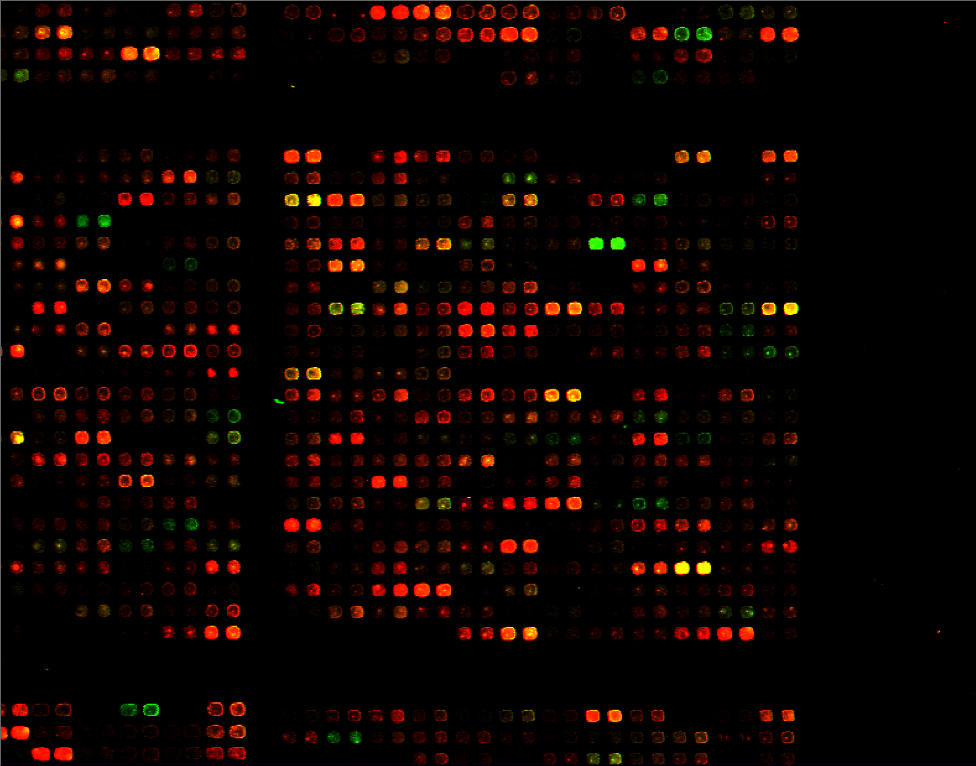
 Click
to see a larger version of this image.
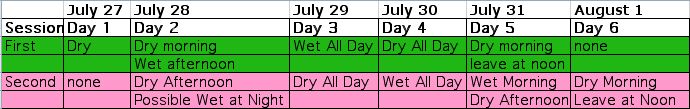
Click
to see a larger version of this image. Two GCAT Best Practices Workshops
July 27 - July 31 and July 28 - August 1
Atlanta, GA @ Morehouse College

Instructors:
Todd Eckdahl, Anne Rosenwald, Mary Lee Ledbetter,
Laurie Heyer,
Laura Hoopes, Malcolm Campbell,
Edison Fowlks
Highlights:
Computers Provided but You Can Bring Laptops
Free Software Available with Training
Produce cDNA probes; Hybe; Wash; Scan; Analyze
Yeast full-genome microarrays
Selection Criteria
Maximize Impact on African American, Hispanic, and Native American Students
Previous Experience with Molecular Methods
Required
(e.g. pipetting, DNA manipulations, etc.)
Desire to Bring Microarrays into Undergraduate Curriculum
Minimal Experience with Microarrays Preferred
Willingness to Learn and Collaborate with Other Faculty
Applied for 2004 Workshop but Not Accepted
(please include this information on your application)
Time of Application Submission
(used as tie
breaker)
Purpose: Genome Consortium for Active Teaching (GCAT) offers hands-on workshops for faculty who conduct research with undergraduate students and/or teach undergraduate courses to learn about gene expression analysis via microarrays.
Workshop Organization:
Part 1 of the workshop
begins with data
analysis (see schedule below)
to introduce the microarray method and will cover data analysis using
public domain data from the literature and from GCAT. Participants
will learn public domain and open source MAGIC
Tool spot-finding and analysis software. The MIAME background information
and its use in microarray evaluation will be covered. Participants
will analyze data sets in short projects. Each of the two workshops
has room for 24 participants.
Part 2 will be a hybridization workshop which will involve hands-on preparation of fluorescently labeled probes for yeast expression microarrays, their hybridization, data acquisition and data analysis using the methods presented in part 1 of the workshop.
Part 3 concludes with data analysis of the microarrays you produced. This will also serve to reinforce what you learned about data analysis.
Where: Morehouse College is the host institution for this workshop; participants may stay in air conditioned residence halls and eat meals in a dining hall. Alternatively, participants living nearby may drive to campus.
When: Workshop #1 will be July 27 - July 31. Workshop #2 will be July 28 - August 1.
Costs and Scholarships: The NSF grant supporting this workshop is paying for all expenses (housing, food, lab fees) for each participant with one catch. You pay for your airfare in advance but you will be reimbursed if you submit an action plan for implementing microarrays in your curriculum and submit receipts. For those with financial need for airfare in advance, you may petition to have this paid in advance.
Download Workshop Announcement |
Download Workshop Application |
|---|---|
Email Edison Fowlks or Malcolm Campbell with Questions
© Copyright 2005 Department of Biology, Davidson College,
Davidson, NC 28035
Send comments, questions, and suggestions to: macampbell@davidson.edu