Purpose
In this webpage, I will try to present information on two different yeast genes: DID4 and YKR005C.
DID4: An Annotated Yeast Gene
Location of DID4
DID4 is spans region 437415 to 43818 of chromosome 11 of Saccharomyces cerevisiae, commonly known as brewers or bakerís yeast (Fig 1). In addition an intron of 68 bp separates DID4ís two exons. Unspliced, the gene is 767 bp long. (GeneDB, 2006; <http://www.genedb.org/genedb/Search?organism=cerevisiae&name=DID4>)
Figure 1. Location of DID4 (scientific name YKL002W). (SGD, 2006; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YKL002W>)
What does DID4 do?
DID4 is needed for endocytosis. In particular, it encodes a protein that is part of the ESCRT-III, an endosomal sorting complex required for complex budding of cellular vesicles into late endosomes (SGD, 2006; http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YKL002W) (WormBase, 2006; <http://www.wormbase.org/db/ontology/gene?name=GO%3A0000815;class=GO_term>), (Foundation for Retrovirology and Human Health, 2004; <http://www.aegis.org/conferences/croi/2004/64.html>). Genes, such as DID4, that allow late endosomes to mature, are called class E vacuolar protein sorting (Vps) factors. The Vps pathway is required for the transport of proteins from the Golgi to the vacuole (Eguez, 2000; <http://www.yeastgenome.org/community/meetings/yeast00/abshtml/134.html>).
Interestingly, DID4 did not receive its name directly from investigations focused on endocytosis, but rather from those focused on ubiquitin-protease pathways. One of the functions of ubiquitin, a protein ubiquitous in cells, is to bind specific proteins in order to help the cell degrade these proteins. The Saccharomyces cerevisiae DOA4 gene (Doa4) aids in this process by producing an enzyme that removes ubiqutin from the degradation products and thereby accelerates this protein degradation process. Certain mutations in DID4, as well as in six other genes, suppress the expression of Doa4. The name DID4 was given to represent the fourth cloned Doa4-Independent Degradation gene. Surprisingly all six cloned genes are class E Vps. (Amerik et al. 2000; <http://www.molbiolcell.org/cgi/content/full/11/10/3365>).
Aliases, protein interactions, and mutant phenotypes:
There are at least six other names that the DID4 gene has gone by: GRD7, REN1, VPL2, VPT14, VPS2, CHM2, and its scientific name YKL002W (iHOP, 2006; <http://www.ihop-net.org/UniPub/iHOP/gs/34086.html>). Perhaps DID4 has been named so many times because the gene is involved in many different reactions. At present, DID4 is known to interact with at least 40 other proteins. Of these forty, some are mitochondrial kinases, some are membrane proteins (including those on the Golgi, endoplasmic reticulum and nucleus), some are proteins associated with microtubules, and some are proteins used for fermentation (BIOGRID, 2006; <http://www.thebiogrid.org/SearchResults/summary/34131>).
Because DID4 is involved in so many processes, researchers have found that mutations in this gene result in a wide variety of phenotypes. In trying to identify mitochondrial proteins, Steinments et al discovered a systematic mutation in DID4 that caused a growth defect in yeast growing on a non-fermentable carbon source (2002; <http://www.nature.com/ng/journal/v31/n4/full/ng929.html>). In addition a null mutation in DID4 suppressed defects associated with the loss of Doa4 and caused abnormal secretion of vacuolar proteins, temperature sensitivity and canavanine-hypersensitivity (Amerik et al., 2000; <http://www.molbiolcell.org/cgi/content/full/11/10/3365>). While investigating proteins responsible for glycogen accumulation, Wilson et al. discovered that a homozygous systematic deletion in DID4 decreased glycogen accumulation in yeast cells (2002; <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=12096123>). Finally, while performing genome wide functional analyses, Xie et al discovered a systematic mutation in DID4 which decreased a cellís resistance to rapamycin (2005, <http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=15883373>).
For more information on mutant phenotypes, see SDGís DID4 website, at <http://db.yeastgenome.org/cgi-bin/phenotype/phenotype.pl?dbid=S000001485>
Orthologs:
Orthologs of DID4 are speculated to exist in 33 different species including C. elegans, D. melanogaster, X. tropicalis, T. rupribes, T. nigroviridis, H. sapiens, M. musculus, R. norvegicus, B.taurus, and C. familiaris (Germline, 2006; <http://www.germonline.org/Saccharomyces_cerevisiae/geneview?gene=YKL002W>).
In mice, the
Protein Expressed:
1 mslfewvfgk nvtpqerlkk nqralertqr elerekrkle lqdkklvsei kksakngqva
61 aakvqakdlv rtrnyiqkfd nmkaqlqais lriqavrssd qmtrsmseat gllagmnrtm
121 nlpqlqrism efekqsdlmg qrqefmdeai dnvmgdevde deeadeivnk vldeigvdln
181 sqlqstpqnl vsnapiaeta mgipepigag sefhgnpddd lqarlntlkk qt
(NCBI, 2006; <http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=9755336>).
GO definitions for DID4:
Molecular Process: protein binding
Biological Process:
a) late endosome vacuole transport
b) protein retention in the Golgi apparatus
c) ubiquitin-dependent protein catabolism via the multivesicular
Cellular Component:
a) ESCRT III complex
b) cytoplasm (UniProt, 2006;<http://www.ebi.uniprot.org/entry/P36108>).
YKR005C: an Unknown gene
Near DID4, is a open reading frame (ORF) which is transcribed in the opposite direction (Fig 2). This other gene is currently uncharacterized. What information about the possible protein expressed can one derive simply from knowledge of this gene's position in the genome and the amino acid and nucleic sequences?
,
Position:
Figure 2, shows the position of YKR005C compared to other genes in the area. YKR005C is transcribed in the direction opposite that of DID4. This difference in transcription raises the question whether the proteins produced by these two genes have similar functions. YKR005C is transcribed in the same direction as two other nearby genes, YKL001C, an andenylyl sulfate kinase required for sulfate assimilation and involved in methionine metabolism, and YKR009C, a multifunctional enzyme of the peroxisomal fatty acid beta-oxidation pathway (SGD, 2006; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=MET14>) (SGD, 2006; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=FOX2>). Because both genes are required for metabolism and because genes in the same region and encoded in the same direction many times have similar functions, I suspect that YKR005C is involved in Saccharomyces cerevisiae metabolism.

Figure 2. Segment of the Saccharomyces annotated genome. Verified gene, DID4, and uncharacterized gene, YKR005C, are highlighted in red. (SGD, 2006; <http://db.yeastgenome.org/cgi-bin/gbrowse/scgenome/?name=ChrXI:427421..448187;label=Landmarks%3Aoverview-Everything-Regulatory>)
Analyses of the Amino Acid Sequence.
A. The sequence (protein translation of the coding sequence)
1 mslfewvfgk nvtpqerlkk nqralertqr elerekrkle lqdkklvsei kksakngqv
61 aakvqakdlv rtrnyiqkfd nmkaqlqais lriqavrssd qmtrsmseat gllagmnrtm
121 nlpqlqrism efekqsdlmg qrqefmdeai dnvmgdevde deeadeivnk vldeigvdl
181 sqlqstpqnl vsnapiaeta mgipepigag sefhgnpddd lqarlntlkk qt
(NCBI, 2006; <http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=9755336>).
B. Kyte-Doolittle Hydropathy Plot:
Using the above sequence, I created a Kyte-Doolittle hydropathy plot (Fig 3). From this data, I see that it is unlikely that this protein has a transmembrane region. Though one region of the amino acid sequence is shown to be a possible transmembrane region, it falls on the ends of the sequence. It is significant that the possible transmembrane region is located at the end of the protein because many non-membrane proteins contain a starting transmembrane region which facilitates movement in the Golgi and other structures.

Figure 3. Query results for amino acid sequence using the Kyte-Doolittle hydropathy plot. Note that only one region may possibly be a transmembrane region and that it falls at the end of this sequence.
C. PREDATOR
Using PREDATOR, I found the secondary structure of this amino acid sequence predicted by the algorithm (Fig 4). Though the secondary prediction obtained by PREDATOR is all but certain because proteins fold into their final form during translation and with guidance of chaperone proteins, it seems odd that the only significant secondary structures are alpha helixes. Of the other predicted secondary structures, extended strands are zigzag structures, and random coils are not a particular shape but a statistical distributions of many shapes. In contrast to these forms, alpha helixes play crucial roles in a protein's overall function. For example, protein channels, such as the CF protein, use alpha helixes to create holes in the cellular membrane though which ionic compounds can pass. In addition, Alpha helixes are extremely common for proteins that undergo allosteric modulations. Flexible springs, alpha helixes can be made more or less ridgid through intermolecular interactions. This change in rigidity in turn can cause the protein to dramatically change its shape.
Interestingly, in the secondary structure predicted, the alpha helixes vary in length and are dispersed throughout the sequence. Because the helixes are not of the same length and are not clustered in one section of the sequence, I am more inclined to believe that they allow allosteric interactions to occur in this protein rather than create a channel for specific molecules to pass through.


Figure 4. Secondary structure of amino acid sequence translated from the coding sequence of YKR005C as predicted by PREDATOR, data width= dssp, with the percentage of each type of secondary structure shown below. Note that the only structures that appear in the sequence are alpha helixes, extended strands, and random coils. In addition note how the alpha helix structures are of different lengths and dispersed throughout the structure.
D. Blastp
I performed a Blastp search for the amino acid sequence on NCBIís database. I was unable to find any hits similar to this amino acid sequence. (Fig 5).
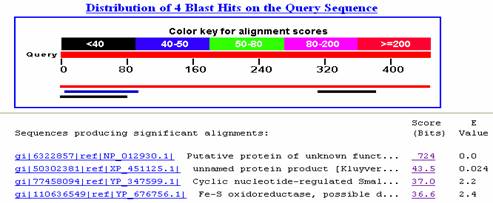
Figure 5. Blastp hits on the protein sequence translated from the coding sequence of YKR005C.
Analysis of the Nucleic Acid Sequence:
A. The coding sequence:
ATGGAGGACTTGGATAACTGCAAATGTTTACAGTGTGCCTTGAGTTCCCTTAATAATAGC
TGCTTTCACGATTACTGTACCTCCAAAGATGAGTATGACAAGTTGCAGATCGTTGTTGAA
CAATTTCAATTGACAAATGGAGTTTTAGATGACGGAGAGATTTTGAAACCTAGAGGCAAC
AAATTTTCTAGTAGAAAATTGTCTTATTTCGTTGGTCAAAACAATACTCTTTTTAGGAAC
CCTTTGCAGTTCGAAAAAAACCAATTAATTTCTGCACTATTAACCTCTTTAACCAATAAT
CAAAAGACTATATCATCAGTTGACATGTTCGAAGTGGTTGACGCTAATAATGAAGTTCAA
TATTTGCGCGAAAGAACGATAAGTGGTAAAACTCTGTCTCCAGCTACTGGTTATGAAGAA
GAAAACGACGGTGACTGTAGTGTGAAAGATAAGAAGTGGGAAGGTAAAATCGAATACCAT
GAGAACAAGAAAGTTAGTTCTGAAAACTGTAGTAAGGATACCGATGATAAAAGTGGCAGT
AAAAAAGAACGAAATACGAAGGCCCCATTATTTCACACTGCCACCGAAATTCATATGACA
AGGTGGAGTTCTTGGAGGCCAAAGAAAATTTTTACTCGTTACTTAGTGAATGAATACCAA
AGTCCTAAAATAATAACCACAGTGAACAGGTTTTACAGGACCAAGACTGATACTGAAACA
GGGACGACTTTAATCACGTCGACAAAAGCCAAAAGAAGATGGTTTCCAAGAACTAAAATC
GTAACAAGTACCGCAACTTCGACGTTTTTGAGTATCACGACTACTACGACAACAAATGCT
ATCGCAACGAAGTCGTTAGTAGCAGTACTTAATCCGGATGGATTGAATAAAAAAGCGGGA
ATAAATTTTGGTCTCTTTAGCGCTAATGGTGAGCTCGCTTCACCAGATGAAGGAGGTACA
CCAACAGTAGTAAGAAGAGATAAGATTTCTGATCCAGGCGCTGCTAATGAGCAGGCCACA
CTGTTCTCTACAACCTTTTCGCAAGTGCCGCACTTACCAGAACTAGATTCTGGTGAATTC
ATATCGGCGGCATCTCAATTAGACAAAAGGATTTTCATATTCACTGCCATCACAGTATCA
ATTACTACATTGATGATGTTGGGGTTTTCTTACCGGTCGCGGGTATCTTTTAGAGATCAT
AGTATCGATGACTCAGATGATGATAACGATTGGTCAGATGACGAAGTTGAATTTGACGAA
GAGTATTTCTATTCTCTTCCTGTAAGCATACCAGAAAAGGGGATCAGCTTAGATAAAATG
GCGCAACAACTTGGTGTAGAATAG
(SDG,
2006;
<http://db.yeastgenome.org/cgi-bin/getSeq>).
B. The genomic sequence:
ATGGAGGACTTGGATAACTGCAAATGTTTACAGTGTGCCTTGAGTTCCCTTAATAATAGC
TGCTTTCACGATTACTGTACCTCCAAAGATGAGTATGACAAGTTGCAGATCGTTGTTGAA
CAATTTCAATTGACAAATGGAGTTTTAGATGACGGAGAGATTTTGAAACCTAGAGGCAAC
AAATTTTCTAGTAGAAAATTGTCTTATTTCGTTGGTCAAAACAATACTCTTTTTAGGAAC
CCTTTGCAGTTCGAAAAAAACCAATTAATTTCTGCACTATTAACCTCTTTAACCAATAAT
CAAAAGACTATATCATCAGTTGACATGTTCGAAGTGGTTGACGCTAATAATGAAGTTCAA
TATTTGCGCGAAAGAACGATAAGTGGTAAAACTCTGTCTCCAGCTACTGGTTATGAAGAA
GAAAACGACGGTGACTGTAGTGTGAAAGATAAGAAGTGGGAAGGTAAAATCGAATACCAT
GAGAACAAGAAAGTTAGTTCTGAAAACTGTAGTAAGGATACCGATGATAAAAGTGGCAGT
AAAAAAGAACGAAATACGAAGGCCCCATTATTTCACACTGCCACCGAAATTCATATGACA
AGGTGGAGTTCTTGGAGGCCAAAGAAAATTTTTACTCGTTACTTAGTGAATGAATACCAA
AGTCCTAAAATAATAACCACAGTGAACAGGTTTTACAGGACCAAGACTGATACTGAAACA
GGGACGACTTTAATCACGTCGACAAAAGCCAAAAGAAGATGGTTTCCAAGAACTAAAATC
GTAACAAGTACCGCAACTTCGACGTTTTTGAGTATCACGACTACTACGACAACAAATGCT
ATCGCAACGAAGTCGTTAGTAGCAGTACTTAATCCGGATGGATTGAATAAAAAAGCGGGA
ATAAATTTTGGTCTCTTTAGCGCTAATGGTGAGCTCGCTTCACCAGATGAAGGAGGTACA
CCAACAGTAGTAAGAAGAGATAAGATTTCTGATCCAGGCGCTGCTAATGAGCAGGCCACA
CTGTTCTCTACAACCTTTTCGCAAGTGCCGCACTTACCAGAACTAGATTCTGGTGAATTC
ATATCGGCGGCATCTCAATTAGACAAAAGGATTTTCATATTCACTGCCATCACAGTATCA
ATTACTACATTGATGATGTTGGGGTTTTCTTACCGGTCGCGGGTATCTTTTAGAGATCAT
AGTATCGATGACTCAGATGATGATAACGATTGGTCAGATGACGAAGTTGAATTTGACGAA
GAGTATTTCTATTCTCTTCCTGTAAGCATACCAGAAAAGGGGATCAGCTTAGATAAAATG
GCGCAACAACTTGGTGTAGAATAG
(SDG, 2006; <http://db.yeastgenome.org/cgi-bin/getSeq>).
C. Analyses of these two nucleic acid sequences (Blastn):
I performed a blastn search for the coding sequence on the NCBI database (Fig 6). The results show that there is little similarity between this gene and others on the NCBI gene database.

Figure 6. Blastn hits for the coding sequence of YKR005C on the NCBI database. Note that in species other than S. cerevisiae, expected values are high.
I also performed a blastn search for the genomic sequence on the NCBI database (Fig 7). Again, the results show that there is little similarity between this gene and others on the NCBI gene database.

Figure 7. Blastn hits for the genomic sequence of YKR005C on the NCBI database. Note that in species other than S. cerevisiae, expected values are high.
Summary
In conclusion, by considering YKR005Cís position on chromosome 11, I believe that YKR005C encodes a protein primarily used during metabolism. In addition, by looking at a Kyte-Doolittle hydropathy plot, I believe that this gene is more likely found in the cytoplasm than attached to a membrane. From looking at its secondary structure as predicted by PREDITOR, I believe that this proteinís function is greatly dependent on allosteric interactions. Finally by conducting Blast searches on the amino acid and nucleic sequences, I find that there is little similarity between this gene and genes found in other organisms. Based on these findings, I believe that YKR005C is an enzyme that regulates metabolic processes unique to S. cervisiae.
Bibliography:
Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. (2000) The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell 11(10):3365-80. <http://www.molbiolcell.org/cgi/content/full/11/10/3365> Accessed 2006 Oct 3.
[BIOGRID] General Repository for Interaction Datasets. 2006. DID4. <http://www.thebiogrid.org/SearchResults/summary/34131>. Accessed 2006 Oct 4.