
STM1/YRL150W and YLR152C: My Favorite Yeast Genes
This web page was produced as an assignment for an undergraduate course at Davidson College.

(SGD 2006; http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000004140)
Note: Except for the conclusions and predicted gene ontology sections, all information for STM1 came from (SGD 2006 A) and all information for YLR152C came from (SGD 2006 B) unless stated otherwise in the text.
My reasoning
While searching through the known and annotated yeast genes, I found a gene that had a biological process called anti-apoptosis. Right away I knew that I wanted to learn more about STM1, a gene that helps prevent cell suicide. Once I had found STM1, I walked along the chromosomal map until I found YLR152C. While this ORF is uncharacterized, people do know that it is not an essential gene. I decided that not essential does not mean not interesting and decided to see what I could find on this ORF. The information that follows will provide insight into how the two genes function in the cell.
STM1
Name
Systematic Name: YRL150W
Alias: MPT4
Interesting fact: STM1 stands for suppressor of tom1. Occasionally TIF3, a translation initiation factor, has been referred to as STM1 as well. All information on this page has been checked to ensure that all information truly refers to the anti-apoptosis gene STM1 and not the translation initiation factor.
Location
Chromosome 12 base 440468 to 441289
(SGD 2006; http://db.yeastgenome.org/cgi-bin/locus.pl?locus=MPT4)
Sequences
Gene Ontology
Molecular Function
- DNA Binding
- Telomeric DNA Binding
Especially binds purine motif triplex and G4 quadruplex in DNA and RNA
Biological Process
- Anti-apoptosis
- Telomere Maintenance
Cellular Component
- Cytoplasm
- Cytosolic ribosome
- Nuclear telomere cap complex
The Kyte-Doolittle Plot below based on the translated coding sequence supports the listed cellular component of STM1 because it lacks peaks above the line that would indicate transmembrane regions. (Johnson S, et al 2006 http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm)

Mutants
- Systematic deletion results in a still viable organism. This mutant was characterized by Giaever G, et al in 2002.
- Null mutation results in a viable organism. When hydrogen peroxide is introduced, null mutants do not experience or experience decreased apoptosis-like cell death. Based on the molecular function and biological processes that STM1 is related ( see information above), I do not know how decreased cell death in the presence of hydrogen peroxide is related to STM1 not functioning. Because this phenotype has been observed, I suspect that there is some function or process that STM1 is a part of that we have not found or linked to STM1.
- Mutant that causes overexpression results in the suppression of some pop2 null mutants', tom1 mutants', and htr1 mutants' phenotypes.
- There are no recorded single nucleotide polymorphisms (SNPs) for this gene (NCBI 2006).
More possible functions
While the gene ontology listing of SGD only provides the functions and biological processes above, a 2004 article by Van Dyke M, et al. suggested that more functions are possibly linked to STM1. The paper notes that while it is found in the nucleus, preferentially binds to telomeres, and has been documented to interact with MEC3 (a protein responsible for telomere silencing, maintenance, and DNA damage check) and Cdc13 (a protein responsible for telomere replication and integrity), it is found in large quantities in the cytoplasm attached to ribosomal proteins and rRNA. It also suppresses temperature-sensitive tom1 and htr1 mutants, staurosporine-sensitive pop2, ccr4, and pkc1 mutants, and a caffeine-sensitive mpt5 mutant, which have no known relation to telomeres or ribosomes. Thus, it is proposed that STM1 either plays a role in multiple processes or it plays a role in transcription or translation, which in turn affects multiple more specific processes (Van Dyke M et al. 2004).
Conserved Domains
No conserved domains were found using NCBI.
Homologs and BLAST
STM1 has three homologs according to NCBI HomoloGene. Of the listed homologous genes, the homolog AGL151W in E. gossypii is the only one that actually is known to code a protein but no experimental data exists for it. MG05307.4 in M. grisea is listed as having a hypothetical protein while the entry for GeneID:2704346 in N. crassa has been removed because a new gene assembly has taken the place of the original shotgun sequence (NCBI HomoloGene (A) 2006).
The NCBI BLASTn results for the full sequence of STM1 returned 2 unique sequences from other organisms that had good E-values as well as several different entries that contained yeast STM1 either by itself or with the surrounding genes. The coding sequence produced similar results. From the two unique sequences (Candida glabrata CBS138, CAGL0E00869g partial mRNA and Kluyveromyces lactis strain NRRL Y-1140 chromosome D), Candida glabrata has an affinity for "guanine-rich nucleic acids" similar to STM1. The sections that are conserved between the two sequences probably relate to this affinity.
The NCBI BLASTp results for the translated coding sequence returns the results expected from the nucleotide but also returns the protein Ray38p in Candida maltosa, which lists the STM1 protein as a homolog. Other hypothetical proteins as well as known proteins also return decent E-values. Hits with an E-value less than 1 are listed below.

Predicted Secondary structure
The predicted secondary structure of the protein contains four sections of alpha helices and no beta sheeting; thus, the protein probably lacks rigidity and is more flexible.
blue = alpha helix
purple = random coil
red = extended stranded
(PBIL 2005; http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html)
STM1 Conclusions
STM1 is most likely involved in many different specific biological processes as witnessed by its presence in both the cytoplasm and nucleus, and its interactions with other proteins, DNA, and rRNA (Van Dyke M, et al. 2004). The most prominently recognized functions seem to be telomere maintenance and anti-apoptosis. The predicted structures lack of beta sheeting points to a more flexible structure that could possibly allow for the protein to associate with all of the different molecules that it interacts with. Finally, the homologs and BLAST hits found in other organisms hint that this is not just an anomaly in the yeast genome.
YLR152C
Name
Standard: None
Systematic: YLR152C
Location
Chromosome 12 base 444689 to 442959
(SGD 2006; http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YLR152C)
Sequences
Gene Ontology
Molecular Function
- unknown
Biological Process
- unknown
Cellular Component
- unknown
Mutants
- Systematic deletion mutant is viable.
- There are no recorded SNPs for this ORF (NCBI 2006)
Conserved Domains
- COG0679: Predicted permeases [General function prediction only]
- pfam03547.10 Auxin_eff: Auxin Efflux Carrier. This family of transporters are found in all domains of life. (There are two of these regions)

(NCBI Conserved Domains 2006; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?INPUT_TYPE=precalc&SEQUENCE=6323181)
Homologs and BLAST
HomoloGene returned 1 homolog, KLLA0D17314g in K. lactis. The gene produces an unnamed protein, which contains the same two conserved domains as YLR152C (NCBI HomoloGene (B) 2006). NCBI's Gene database uncovered AGL149C in Eremothecium gossypii, which produces a protein that is a "syntenic homolog of Saccharomyces cerevisiae YLR152C" but only contains the two Auxin_eff conserved domains (NCBI Gene 2006).
NCBI BLASTn of both the full and coding sequences returned poor results. The closest hit was Zygosaccharomyces bailii, which contained a partial YLR152C gene.
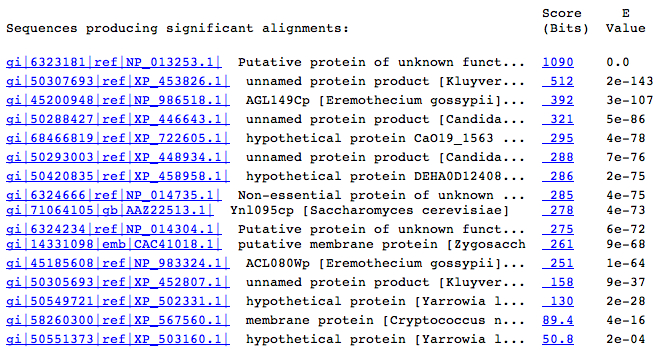
NCBI BLASTp of the translated ORF coding sequence returned many results, but after the first two that were not YLR152C, the identities dropped below 40% even though the E-values remained very low. The conserved domains in YLR152C are responsible for the large number of hits. Many of the proteins, both hypothetical and confirmed, are identified as membrane proteins. Below is a list of all hits that had E-values less than .001.
Kyte-Doolittle Hydropathy Plot
The four peaks in the Kyte-Doolittle plot predict that YLR152C contains four transmembrane regions (Johnson S, et al 2006; http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm).

Predicted Secondary structure
The predicted secondary structure contains 14 alpha helix regions (29.34% of the structure) while the rest of the structure is extended strand and random coil. Because of the large number of alpha helices and complete absence of beta sheets, the protein should be rather flexible.
blue = alpha helix
purple = random coil
red = extended stranded
(PBIL 2005; http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html)
Predicted Gene Ontology
Molecular Function
- efflux permease activity
Biological Process
- transport
Cellular Component
- plasma membrane
YLR152C Conclusions
While definite information on YLR152C is extremely sparse and rare compared with an annotated gene like STM1, a tentative prediction of gene ontology can be made using information from conserved domains, BLAST searches, and programs that generate predicted structures and features based on translated coding sequences. The Kyte-Doolittle plot predicted transmembrane regions in the protein; thus, the information helped support the presence of conserved domain regions that often signify membrane channels. The BLAST hits that were known proteins also hint at possibile locations and functions. The flexible structure predicted by Predator can also be used to support the claim that it is a channel protein. Channel proteins often change configuration to allow the proper molecule to pass through before blocking off entrance into the cell once more. The alpha helices in the secondary structure suggest that this protein would be capable of the configuration change. Although wet lab work is needed to back up all of these predictions like in the annotated gene, we can at least form an idea of where to start investigating in the lab.
References
SGD References
A. SGD. 2006 Oct. STM1/YRL150W Summary.<http://db.yeastgenome.org/cgi-bin/locus.pl?locus=MPT4>.Accessed 2006 Oct. 1.
B. SGD. 2006 Oct. YLR152C Summary.<http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YLR152C>.Accessed 2006 Oct. 1.
NCBI References
NCBI. 2006 Oct. 5. < http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi>. Accessed 2006 Oct. 1-6.
NCBI Conserved Domains. 2006 Oct. 5. Query sequence: [gi|6323181|ref|NP_013253.1|] <http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?INPUT_TYPE=precalc&SEQUENCE=6323181>. Accessed 2006 Oct. 4.
NCBI Gene. 2006 July 7. AGL149C AGL149Cp [ Eremothecium gossypii ]. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene&cmd=Retrieve&dopt=full_report&list_uids=2758374 >. Accessed 2006 Oct. 4.
NCBI Homologene (A). 2006 Oct. 2. HomoloGene:39708. Gene conserved in Ascomycota http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=homologene&dopt=HomoloGene&list_uids=39708>. Accessed 2006 Oct. 4.
NCBI Homologene (B). 2006 Oct. 2. HomoloGene:39710. Gene conserved in Saccharomycetaceae. <http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?INPUT_TYPE=precalc&SEQUENCE=6323181>.Accessed 2006 Oct. 4.
Other References
Johnson S, McCord RP, Robinson L, and Heyer L. Kyte-Doolittle Hydropathy Plot. <http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm>. Accessed 2006 Oct. 3.
PBIL. 2005 Oct. 20. Predator Secondary Structure Prediction Method. <http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html>. Accessed 2006 Oct. 3.
Van Dyke MW, Nelson LD, Weilbaecher RG, and Mehta DV. 2004 June 4.Stm1p, a G4 Quadruplex and Purine Motif Triplex Nucleic Acid-binding Protein, Interacts with Ribosomes and Subtelomeric Y' DNA in Saccharomyces cerevisiae* In J. Biol. Chem. 279(23): 24323-24333. JBC online <http://www.jbc.org/cgi/content/full/279/23/24323 >. Accessed 2006 October 5.
Links
Erin's Genomics Web page
Genomics Course Page
Biology Home PageThis page was created by Erin Zwack. Comments and questions are welcome.