
This web page was produced as an assignment for an undergraduate course at Davidson College.
Sequencing the Genomes of Ancient Canids

The following is a review of the article "Complete Mitochondrial Genomes of Ancient Canids Suggest a European Origin of Domestic Dogs" by Thalmann et al. A full citation and link to the article can be found at the bottom of this webpage. All quotations, facts and data presented in this review are drawn from this article unless otherwise cited.
Summary
There is a reason that dogs are known as man’s best friend. Evidence of dog domestication exists from as long as 36,000 years ago, implying a long and rich relationship between man and dog. However, tracing the evolutionary origins of dog is hindered by human manipulation of the canine genome via selective breeding and a long history of trading different breeds of dogs between far reaching corners of the world. Thalmann et al. sought to navigate this complicated task and create a phylogenetic tree relating modern day dog breeds to DNA derived from ancient canids and modern day wolves.
Thalmann et al. sequenced mitochondrial DNA obtained from 18 of the earliest “wolflike” and “doglike” canid species (Table 1). They compared this data to mtDNA drawn from 49 modern day wolves , a wide variety of modern day dog breeds totaling 77, and four coyotes. From this data, the authors constructed a phylogenetic tree that grouped the modern day dog species into four distinct clades (Figure 1).
Of these four clades, Clade A is the most phylogenetically diverse, containing 64% of the dog sequences. Its closest ancient relatives are two pre-Columbian new world dog species; the two groups share a most recent common ancestor ~18,800 years ago. These pre-Columban New World dogs and Clade A are most closely related to an ancient wolf canid found in Switzerland, dating to around 14,000 years ago.
Clades B and C, which contain 22 and 12 % of the dog sequences respectively, are also closely related to European sequences. Clade B is most closely linked to modern wolves in Sweden and Ukraine, with who it shares a most recent common ancestor ~9200 years ago. Clade C shares a common ancestor with an ancient canid species found in a cave in Germany.
Clade D, the clade with the lowest diversity of modern day dog species, contains just 2% of the dog DNA sequenced. The sequences contained in Clade D include those from 2 Scandinavian breeds, who appear to be a sister group to an ancient canid found in Switzerland. This branch of the phylogenetic tree is rooted in a sequence from an ancient Russian canid sample. From this data, it appears that this is the only clade not rooted in European descent.
Previous research has suggested that dog domestication originated in the Middle East or Asia. Thalmann et al. claim that as the majority of the dog breeds whose DNA they sequenced seem to be of European descent, that dog domestication actually began in Europe rather than the Middle East or Asia. Additionally, “Bayesian analysis of divergent times implies a European origin of the domestic dog dating to as much as 18,800 to 32,100 years ago,” a range consistent with the most recent common ancestors for the four clades rooted in European descent that their data established.
Thalmann et al. also use Bayesian skid analysis to look at the populations of dog Clade A and the pre-Columban dogs. Their results show a steady increase until about 5,000 years ago, which the authors attribute to the domestication of dog. From 5,000 to 2,500 years ago, dog populations experienced a decrease, which was followed by a sharp increase in population size. The authors suggest that, as this parallels what is known of human population sizes during this time, that dog population size is dependent on that of human society.
To recap...
1. Researchers compared modern day dog mtDNA to mtDNA from wolves and ancient canids
2. This data suggests a most recent common ancestor with wolves and ancient canids of European descent, rather than Middle Eastern or Asian as previously suggested.
3. Data collected also supports the hypothesis that dog population density is highly dependent on human population patterns.
Evaluation
The main point of the authors' interpretation of their data is to establish the origins of dog domestication as European rather than Middle Eastern or Asian. However, the researchers fail to include any DNA from any ancient Middle Eastern or Asian canids. This is a major weakness of the study, and really undermines the claim the researchers are trying to make.
The researchers address this shortcoming briefly by saying, “Notably, our ancient panel does not contain specimens from the Middle East or China, two proposed centers of origin. In fact, no ancient dog remains older than ~13,000 years are known from these regions.” While data does indicate that dog domestication occurred some time prior to this point in time, these remains should have been included in the study to provide some sort of data for ancient dogs from these regions. Ancient DNA dating more recently than this was included in the study (both from Argentina and the USA), leaving no valid reason for the exclusion of the Middle Eastern or Chinese canid DNA.
Without any DNA from ancient Asian or Middle Eastern canids, all that this study does is establish that ancient European dogs and modern day dogs are related. Since it does not look at the relationship between ancient Middle Eastern/Asian DNA and modern day dog DNA, claiming to prove a closer evolutionary relationship to European canids seems a bit premature and unsupported.
Another weakness of this study is that they look only at a single locus within the mitochondrial genome. The researchers recognize this weakness, and say that, “given the recent divergence of dogs and wolves” this weakness “can potentially confound evolutionary inverence.” While they acknowledge the need for the examination of more loci to refine their phylogenetic tree, this shortcoming combined with the aforementioned lack of ancient Middle Eastern/Asian canid DNA makes me even more hesitant to accept the conclusions they draw from their data as indisputable.
Additionally, the researchers often make vague statements regarding their methods without providing the background or details necessary to truly evaluate the effectiveness of the study and the validity of the results. How the ancient DNA was obtained, preserved, and validated as actually ancient DNA was largely ignored. This information is critical when dealing with ancient DNA due to the often contamination of so called “ancient DNA” with modern day DNA.
As a whole, I thought that the article was largely disorganized and ineffectively structured. Table 1 and Figure 2, though discussed in the text of the article, did not appear until the very end. Additionally, the discussion of the results lacked any sort of intentional structure, making a fairly straightforward study and results sometimes difficult to follow.
Figures
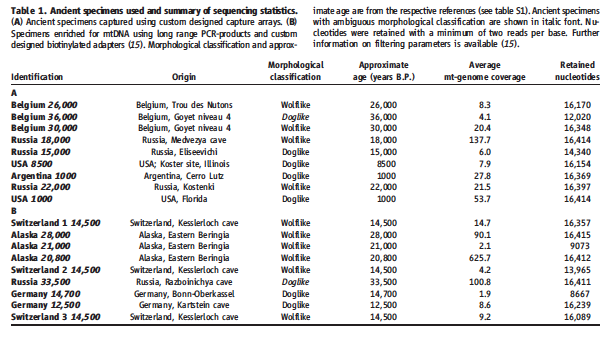
Table 1 summarizes the 18 ancient canid species analyzed in this summary. The different species are separated into groups A and B based on what method of sequencing was used. However, the authors provide no explanation as to what methodology was behind the separation of these two groups. Examination of the different information supplied (nucleotide length, age, and wolf vs. dog like) yields no apparent, consistent trends to the separation.
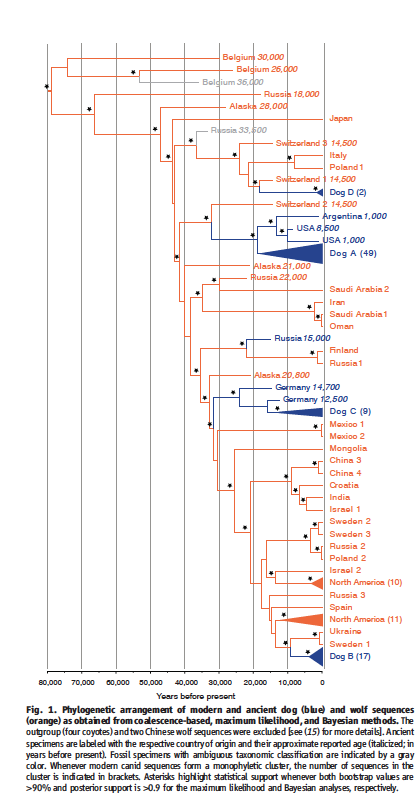
Figure 1 is the phylogenetic tree researchers created using the DNA sequences they collected from ancient canids, modern day wolves, and modern day dogs. The x-axis measures "years before present," making the horizontal axis the relevant measure of evolutionary distance between species. Notable conclusions are discussed in the summary section, but most notably include the close relationships of Clades A, B, and C to ancient European canids.
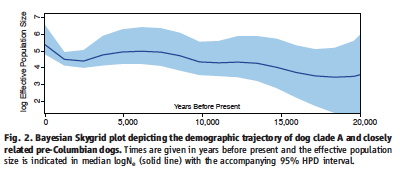
Figure 2 plots the effective population size of dog, as determined by Bayesian methods, against time. Conclusions and inferences are discussed in the summary section, but include the paralleling of dog populations with those of humans. Consequently, it would be useful to have a reference plot of human population size with this graph. Additionally, it would be useful if the researchers had clearly marked specifically discussed points in time for ease of understanding. Finally, the x-axis on this figure seems contrary to common sense. The majority of the world's written languages leads them to read from left to right, a concept that translates to reading scales of time. Therefore, it first seems illogical that the most recent time period is furthest to the left while the most ancient time period displayed on this graph is located all the way to the right. A different x-axis, or a distinct "present day" label could have helped circumvent this problem.
Evaluation of Figures
For the most part, these figures are straightforward and easy to understand. They are not incredibly complex or complicated, but also do not offer any revolutionary insights or help with any deeper understanding of the material.
References
Supplemental Images


Genomics Page © Copyright 2014 Department of Biology, Davidson College,
Davidson, NC 28035



Biology Home Page