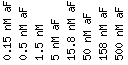Expression of Saccharomyces' ORF YEL068C
YEL068C is an ORF with no known
function (SGD
2001). The analysis YEL068C expression and its comparison to other genes'
expression under certain conditions can provide clues as to the function of the
YEL068C in the Saccharomyces' cell cycle. A search for ORF YEL068C on the
Saccharomyces Genome Database's Expression Connection outlines the expression
data for YEL068C in microarray experiments involving YEL068C's expression
depending on alpha-factor concentration, in
response to alpha-factor, during the diauxic shift, during sporulation, in
response to histone depletion, during the cell cycle, and in response to
glucose limitation in evolved strains.
Each experiment
uses this scale to represent induction or repression:
![]()
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
YEL068C’s response to
increased concentration of alpha-factor
(Roberts
et.al 2000)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
SRY1 |
|
|
serine metabolism |
|
not yet annotated |
|
not yet annotated |
|
|
|
|
UBC8 |
|
|
polyubiquitylation* |
|
ubiquitin conjugating
enzyme |
|
cellular_component unknown |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
ECM13 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
CTK1 |
|
|
protein amino acid
phosphorylation* |
|
protein kinase |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
AUT7 |
|
|
autophagy* |
|
microtubule binding |
|
microtubule associated protein |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
THI5 |
|
|
vitamin B1 biosynthesis |
|
molecular_function unknown |
|
cellular_component unknown |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
MBR1 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
FAD1 |
|
|
not yet annotated |
|
FMN adenylyltransferase |
|
not yet annotated |
This
image was taken from the SGD webpages’s Expression
Connection database.
• This experiment uses varying concentrations alpha- factor,
which is a Saccharomyces' pheromone, to reveal resulting expression among
genes. YEL068C expression increases slightly with increased concentration of
alpha -factor. YEL068C maximum induction occurred at 158 nM aF with 0.25 fold
induction but decreased again at 500 nM aF to approximately 0.1 fold induction
(AC Rosetta, 2001). Genes with similar induction patterns are located on different
chromosomes (GO report, 2001). Many of the genes also have unknown biological
processes, but SRY1, UBC8, CTK1, AUT7, and THI5 are all involved in metabolism.
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
YEL068C’s expression in
response to duration of exposure to alpha-factor (Roberts et.al 2000)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
PEX10 |
|
|
peroxisome organization and
biogenesis |
|
molecular_function unknown |
|
cellular_component unknown |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
PET56 |
|
|
ribosomal large subunit
assembly and maintenance |
|
rRNA
(guanine-N1-)-methyltransferase |
|
mitochondrion |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
PRR2 |
|
|
MAPKKK cascade |
|
receptor signaling protein
serine/threonine kinase |
|
cellular_component unknown |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
GLO4 |
|
|
carbohydrate metabolism |
|
hydroxyacylglutathione
hydrolase |
|
mitochondrion* |
|
|
|
|
CDC9 |
|
|
nucleotide-excision repair* |
|
DNA ligase (ATP) |
|
replication fork |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
MPT1 |
|
|
transcription, from Pol II
promoter |
|
RNA polymerase II transcription
factor |
|
TFIID complex |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
QRI5 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
SPO74 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
YRA1 |
|
|
mRNA processing |
|
not yet annotated |
|
not yet annotated |
|
|
|
|
NUM1 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
|
|
|
|
|
|
|
biological_process unknown |
|
molecular_function |
|
|
This image was taken from the SGD webpages’s Expression Connection database.
• This experiment uses duration
of exposure to alpha-factor to reveal resulting expression among genes. YEL068C
expression was repressed slightly after 90 minutes of exposure to alpha-factor.
The genes with a similar expression pattern to YEL068C include those involved
in DNA repair and replication, mRNA processing, carbohydrate metabolism, MAP
signal transduction pathway, and ribosome assembly and maintenance (GO report,
2001).
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
YEL068C’s expression
during diauxic shift (DiRisi et.al 1997)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SNT309 |
|
|
mRNA splicing |
|
not yet annotated |
|
not yet annotated |
||
|
|
ABF2 |
|
|
mitochondrial genome
maintenance |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
CDD1 |
|
|
cytidine catabolism* |
|
cytidine deaminase |
|
cellular_component unknown |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SSO2 |
|
|
non-selective vesicle
fusion* |
|
t-SNARE |
|
integral plasma membrane
protein |
||
|
|
CET1 |
|
|
not yet annotated |
|
polynucleotide
5'-phosphatase |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
CSE2 |
|
|
transcription, from Pol II
promoter* |
|
RNA polymerase II
transcription mediator |
|
mediator complex |
||
|
|
JNM1 |
|
|
mitotic anaphase B |
|
structural protein of
cytoskeleton |
|
dynactin complex |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SWI3 |
|
|
chromatin modeling |
|
non-specific RNA polymerase
II transcription factor |
|
nucleosome remodeling
complex |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
POS5 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SUB1 |
|
|
transcription |
|
not yet annotated |
|
not yet annotated |
This image was taken from the SGD webpages’s Expression Connection database.
· YEL068C
is repressed at intervals during the time the cell under went the shift from
anaerobic to respiratory metabolism. Many of the cells with similar expression
patterns are not annotated; but SUB1, SWI3, SCSE2, JNM1, and SNT309 are
involved in the cell cycle.
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
YEL068C’s
expression during sporulation (Chu et.al 1998)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
KRE26 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
DCG1 |
|
|
biological_process unknown |
|
not yet annotated |
|
not yet annotated |
||
|
|
YPS5 |
|
|
not yet annotated |
|
aspartic-type endopeptidase |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
HPA3 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SAR1 |
|
|
ER to Golgi transport |
|
SAR small monomeric GTPase |
|
COPII vesicle coat |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
COD5 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
YPT31 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
RNH35 |
|
|
DNA replication |
|
ribonuclease H |
|
cell |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SEN2 |
|
|
tRNA splicing |
|
tRNA-intron endonuclease |
|
nuclear inner membrane* |
This image was taken from the SGD webpages’s Expression Connection database.
· YEL068C
is repressed before two hours of exposure; then it is induced at two hours; and
then it is repressed again for the entire of the duration. Genes with similar
expression patterns to YEL068C during sporulation function in the cell cycle or
in protein synthesis or are unknown.
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
YEL068C’s
expression as histone is depleted (Wyrick
et.al 1999)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SBE2 |
|
|
bud growth |
|
molecular_function unknown |
|
cell |
||
|
|
PSP1 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
HCA4 |
|
|
35S primary transcript
processing |
|
ATP dependent RNA helicase |
|
nucleolus |
||
|
|
BET3 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
OAF1 |
|
|
transcription regulation,
from Pol I promoter |
|
specific RNA polymerase II
transcription factor |
|
nucleus |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
ASH1 |
|
|
pseudohyphal growth* |
|
specific transcriptional
repressor |
|
nucleus |
||
|
|
LOS1 |
|
|
tRNA splicing |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
APS2 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
ENT1 |
|
|
actin filament
organization* |
|
cytoskeletal adaptor |
|
actin cortical patch (sensu
Saccharomyces) |
||
|
|
FKH2 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SIS2 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
KNS1 |
|
|
protein amino acid phosphorylation |
|
protein serine/threonine
kinase* |
|
not yet annotated |
||
|
|
MYO4 |
|
|
mRNA localization, intracellular |
|
microfilament motor |
|
actin cap (sensu
Saccharomyces) |
||
|
|
GPE2 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
This image was taken from the SGD webpages’s Expression Connection database.
YEL068C is repressed for the entire duration
of histone depletion. It has a similar expression pattern to genes involved in
protein synthesis (MYO4, KNS1, LOS1, OAF1, HCA4) and structural genes (ENT1,
ASH1, SBE2).
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
YEL068C’s
Expression during Cell Cycle (Spellman et.al 1999)
|
Name |
Score |
Peak |
|
|
5.047 |
S |
|
|
|
Reference Genes |
|
||
|
10.9 |
G1 |
|
|
|
10.68 |
S |
|
|
|
3.08 |
G2 |
|
|
|
6.726 |
M |
|
|
|
11.8 |
M/G1 |
|
|
|
|
|
|
|
|
Plot of YEL068C |
|
||
This image was taken from the SGD webpages’s Expression Connection database.
· This experiment analyzes the expression of genes during the cell
cycle. YEL068C is repressed and induced during different stages of the cell but
with a little deviance from 1:1 fold expression.
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
YEL068C’s
expression evolution through generations with limitations on glucose supply (Ferea et.al
1999)
|
Name |
|
|
Process |
|
Function |
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
not yet annotated |
|
not yet annotated |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
actin filament
organization* |
|
RAB GTPase activator |
|
|
|
|
not yet annotated |
|
molecular_function unknown |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
mRNA-nucleus export* |
|
structural protein |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
protein-mitochondrial
targeting |
|
molecular_function unknown |
|
|
|
|
transcription |
|
not yet annotated |
|
|
|
|
ubiquitin-dependent protein
degradation* |
|
enzyme activator |
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
|
|
|
protein-peroxisome
targeting |
|
protein binding |
|
|
|
|
G1/S transition of mitotic
cell cycle |
|
protein serine/threonine
phosphatase |
|
|
|
|
mitotic anaphase B* |
|
microtubule binding |
This
image was taken from the SGD webpages’s Expression Connection database.
·This experiment regulates the glucose supply for yeast over
several generations and then test the evolved colonies on a glucose limited
medium to determine evolution of expression of genes. YEL068C is repressed in
the evolved second generation in comparison to the parent generation. Genes
with similar expression patterns are involved in the cell cyle and structure.
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
Although nothing concrete
can be assumed from the data generated in these microarrays about the function
of YEL068C, the fact that it generally clustered with genes involved in cell
cycle regulation, protein synthesis, and structure is very compelling.
Return to Microarray Main Page