
A recent study published in Natureattempted to identify common genetic factors between individuals with ASDs. They began by taking tissue samples from 780 families comprised of 3,101 individuals. Each family contained at least two individuals with an ASD as identified by the Autism Diagnostic Interview-Revised (ADI-R) and Autism Diagnostic Observation Schedule (ADOS).
In a separate study, samples were taken from 1,204 individuals with an ASD and 6,491 who were identified as unaffected by the disease and acted as a control. The team was looking for single nucleotide polymorphisms (SNPs) that individuals with an ASD had in common that were different from the rest of the unaffected population. A SNPis a single nucleotide that varies at a specific locus.The purpose was to try and determine part of the genetic component responsible for ASDs. By finding associations between SNPs and ASD expression, it might be possible to identify which genes are affected by these variations. Even if they could not identify the gene, a genom-wide association with specific variations would enable early detection of autism and its associated disorders.

To begin with, the researchers used the Pedigree Disequilibrium Test to look for common SNPs within affected families. Following this approach, no genome-wide association between specific SNPs and ASD disorders appeared. They then looked to the unrelated and control groups and could find no conclusive results in that pool either.
It was then decided that, due to the nature of ASDs, it would be impossible to locate SNPs with such small data pools. Because ASDs do not present with a single phenotype and have such variable symptoms (and, it is assumed, genotypes), a much larger pool was necessary. The reasearchers then combined the related and unrelated groups and again searched for SNPs common among ASD sufferers but significnatly different from unaffected individuals.
They found one SNP in the 5p14.1 chromosomal region which reached genome-wide significance.As can be seen in Table 2, rs4307059, along with five other SNPs in the same region, had P values below 1x10^-4. Several other promising SNPs were discovered but the 5p14.1 region had, by far, the lowest and most promising P values.

Further investigation showed the 5p14.1 region contained genes CDH10 and CDH9. It has been documented that significant levels of CDH10 but not CDH9 are involved in fetal brain development in the prefrontal cortex (Figure 2). The researchers threw more of their resources into trying to find a connection between the SNPs they had found and the CDH10 gene. It's involvement in cell-cell adhesion and high expression in the pre-frontal cortex highly suggested that it's disruption would cause a significant change. The fact that the prefrontal cortex has long been known to be involved in the development of language skills and social responses made it even more likely that the researchers had found their gene.
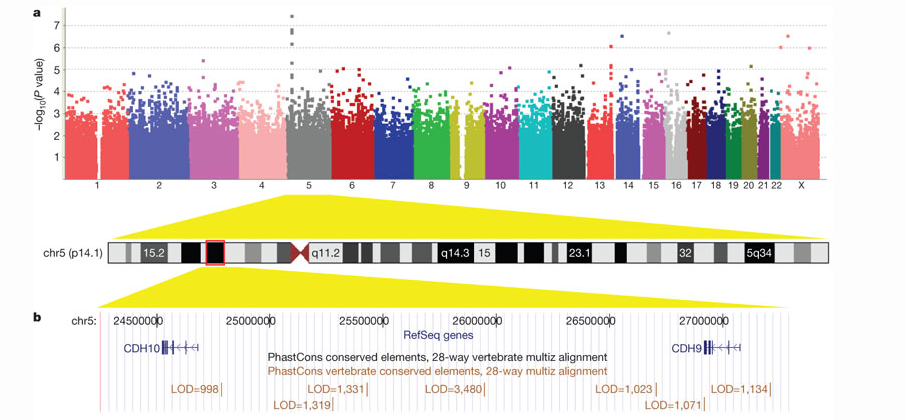
Unfortunately, as can be see in Figure 2, they could not find any significant association between their most promising SNP and CHD10 expression. There was nothing to suggest that the mutations they found would have a functional affect on the CDH10 gene. This does not mean that the study is useless, only that more information needs to be gathered in order to interpret their results.

All from Wang et al, 2009.
This web page was produced as an assignment for an undergraduate course at Davidson College.