This web page was produced as an assignment for an undergraduate course at Davidson College.
Evolution Over Time
Implications for Real Time Clinical Tracking

Klebsiella pneumonia
From planetinvestigations. Currently seeking permission to use this photo.
As whole genome shotgun sequencing becomes even faster, it will likely be possible to track the evolution and spread of pathogens as outbreaks occur. Pathogenomics has already helped track the evolution and spread of disease immediately after an outbreak. For example, in one hospital in Maryland, a particularly virulent form of Klebsiella pneumonia spread. This particular form of the pathogen kills more than half of those it infects (Cossins, 2012). One patient known to have the infection was isolated, but after an unexpected spread to several other patients, researchers sequenced the pathogen genome from infected patients and found several different strains present in infected individuals (Cossins, 2012). By constructing an evolutionary tree using the sequenced genomes, scientists were able to track the evolution and spread of the disease through the hospital (Cossins, 2012). While this application of pathogenomics may not directly identify a form of treatment, it can certainly stop the spread of the pathogen, and prevent infection of more individuals (Cossins, 2012).
Real time whole genome sequencing of clinical samples has huge implications for tracking and treating disease outbreaks in hospital scenarios. Hasman et al note that “rapid turnaround, affordability, and the provision of clinically relevant information to health care personnel that can be interpreted without specialist knowledge of wholegenome sequencing” are all benefits of real time clinical sequencing (Hasman, 2014). They also note that in order for this technology to be useful, the entire process needs to be automated so that clinicians can analyze the data rapidly with little knowledge of the precise methods needed to do so. With clinical application in mind, Hasman et al developed a bioinformatics tool that rapidly analyzes data from clinical samples and identifies “both species and resistance genes in a sample and additionally give[s] information on the presence of other DNA, including human and fungal DNA, all within a clinically relevant time frame” (2014). Further development of this technology combined with increased speed and reduced cost of whole genome sequencing will allow scientists and clinicians alike to track the evolution and spread of pathogens as they occur (in real time), which will in turn allow for swift and effective treatment of pathogen induced diseases.
Burkholderia dolosa
From nationalgeographic. Currently seeking permission to use these photos.
Additional clinical research from Lieberman et al shows the parallel evolution of Burkholderia dolosa, which is a rare cystic fibrosis pathogen, in multiple individuals over a sixteen year period (2012). This study focused on SNPs and noted that SNPs accumulated at a steady rate within all infected individuals (Lieberman, 2012). Through their analysis, Lieberman et al identified seventeen B. dolosa genes that “underwent selective pressure during infection in subjects with cystic fibrosis[, which] highlights key pathways involved in pathogenesis and may suggest new therapeutic targets for this and other lung infections” (2012).
In addition to clinical applications, pathogenomics has epidemiological implications as well. For example, pathogenomics showed that a relatively recent outbreak of cholera in Haiti was a strain that had recently diverged from a strain in Nepal, which allowed scientists to track the spread and evolution of the pathogen (Grad, 2103). This demonstrates a larger scale application for pathogenomics and its role in elucidating the spread and evolution of pathogens.
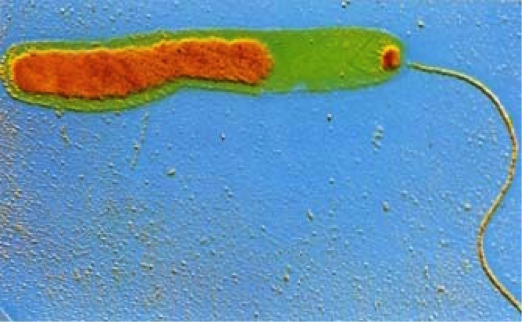
The Vibrio cholerae bacterium under an electron microscope. Color has been added to show the nucleic acid (orange) and the flagellum (tail), which is used by the bacterium to move.
CNRI/Science Photo Library, Photo Researchers, Inc.
From humanillnesses. Currently seeking permission to use these photos.
Resources:
http://www.the-scientist.com/?articles.view/articleNo/33771/title/Real-time-Outbreak-Sequencing/
http://jcm.asm.org/content/52/1/139.long
http://mbio.asm.org/content/4/5/e00670-13.full.pdf+html
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3245322/pdf/nihms335194.pdf
Assignments:
Assignment #2:You get to choose (due Feb. 28, 2 pm)
Assignment #3: You get to choose (due Apr. 25, 2 pm)
Genomics Page
Biology Home Page
© Copyright 2014 Department of Biology, Davidson College, Davidson, NC 28035