This web page was produced as an assignment for an undergraduate course at Davidson College.
Molecular Tool: RNAi
What is RNAi?
RNAi, also known as RNA interference, is a new tool that makes possible the regulation of gene expression after transcription. RNAi works through the introduction of gene- specific dsRNA into a cell, and thus causing the homologous mRNA to be degraded (Cottrell and Doering 2003). RNAi is one of many post- transcriptional gene silencing mechanisms (Ullu and Tschudi 2000). Post- transriptional gene silencing (PTGS) is "the silencing of an endogenous gene caused by the introduction of a homologous dsRNA, transgene, or virus" (Ambion 2002). The transcript of the silenced gene is made by PTGS, but is destroyed quickly after it is made. This results in cosuppression, in which both the introduced gene and the homologous endogenous gene are suppressed (Ambion 2002). Through the use of RNAi, scientists have made some recent improvements in "reverse genetics," that is, they first have the known gene sequence and then work to find its function by inhibiting is activity in vivo (Waksman Student Scholars 2003).
RNAi is believed to be a natural mechanism that acts to protect the DNA of an organism against invading transposons and viruses that produce "aberrant RNA" or dsRNA into the organism's cells (Tuschl 2001). The organisms cells then degrade this RNA to protect themselves from transposon and virus infection through the use of the RNA interference mechanism (Tuschl 2001).
How does RNAi work?
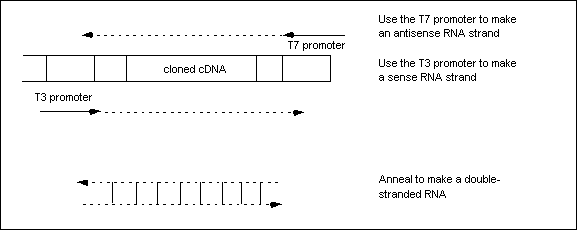
RNA is made in a test tube by adding phage RNA polymerases that bind to phage promoters that are within expression vectors. For example, Phage T7 and T3 promoters can be used within the expression vector with the cDNA of the gene that you wish to express (or in this case, block the expression of) to make double stranded RNA when T7 or T3 polymerases are provided (Waksman Student Scholars 2003). Here, T7 and T3 promoters are on opposite ends of the gene that is to be cloned so that T7 polymerase acts to express the antisense strand, while T3 polymerase acts to express the sense strand (Figure 1) or vice versa (Figure 2).
Figure 1. Figure from Wasksman Student Scholars website, Rutgers University. This figure demonstrates how dsRNA is synthesized in a tube, through the use of T7 and T3 promoters within a phage expression vector. T3 and T7 polymerases bind to this expression vector to express the two strands of dsRNA. Permission Pending. Click on Figure to find out more about source.
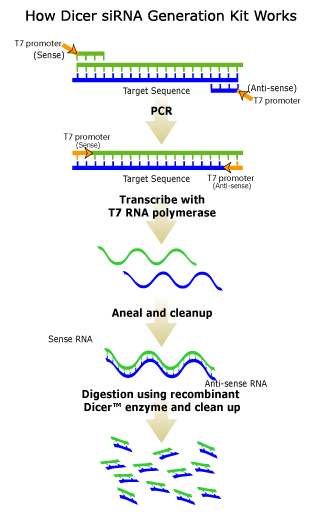
After the cDNA is trascribed through PCR, the sense and antisense strands are annealed together to give double stranded RNA. This dsRNA is then cut into 21-25 by small interfering RNAs (siRNA) by an RNase II, like the Dicer enzyme shown in Figure 2. However, this enzyme also has a helicase section, that is believed to act in the unwinding of the dsRNA that may be necessary for the binding of this RNA to the target RNA that we wish to inhibit the translation of (Shuey, McCallus, and Giordano 2002).
Figure 2. Figure obtained from Gene Therapy Systems, Inc. Through the use of T7 promoters and PCR, dsRNA is synthesized. The enzyme Dicer (a ribonuclease III enzyme) then cleaves this dsRNA into 21-23 nucleotide siRNAs. © 2002 Gene Therapy Systems, Inc. Click on Figure to find out more about the source.
These siRNAs are combined into a complex called the RNA- induced silencing complex (RISC) and then is able to target a specific the cellular RNA for enzymatice degradation (Shuey, McCallus, and Giordano 2002). Some believe that siRNAs are able to degrade their target RNAs by marking it for destruction, which may consist of "covalent modification of the target" by enzymes such as adenosine deaminase or through other mechanisms (Montgomery 2003). It is also believed that one dsRNA could mark many target mRNAs before it is used up (Montgomery 2003). In addition, these siRNAs can be used as primers to make new dsRNA that can then be a target for the enzyme Dicer, and the cycle continues (see Figure 3) (Shuey, McCallus, and Giordano 2002).
Figure 3. This is a figure from Shuey, David J.; McCallus, Daniel E.; and Giordano, Tony. It shows an overview of the RNAi pathway. DsRNA is cut by the enzyme dicer into 21-25 nucleotide siRNAs. SiRNAs then attach to the RNA- induced silencing complex (RISC), and the antisense strand of siRNA is then bound to the target RNA and silence it. SiRNAs are also used as primers for the next generation of dsRNA. This new dsRNA is made by the enzyme RNA polymerase or RdRp. Permission Pending. Click on the Figure to find out more about the source.
Why is RNAi important? How can RNAi be Applied?
As more and more complete genomes are being sequenced, a need to develop the tools for understanding the functions of specific genes, especially open reading frames increases. For this reason, the need for a tool like RNAi is also increasing. In addition, RNAi is especially important in the theraputic applications for a number of diseases (Ullu and Tschudi 2000). There are three main time points at which a disease can be stopped. These are transcriptional, post-transcriptional, and post-translational intervention. Before the discovery of antisense RNA and RNAi, most of the drug targets have been proteins, and therefore, post- translational intervention (Shuey, McCallus, and Giordano 2002). RNAi is a way to control the development of a disease earlier on in the process. The high specificity of RNAi to the target RNA makes it an important mechanism to find out the function of the gene (Tuschl 2001).
Although RNAi has only been shown to attack viruses grown in human cells in the laboratory setting, there has been recent adavancements in the use of RNAi within mammalian cells. It has been shown by Judy Lieberman and collegues at the Harvard Medical School in Boston that RNA interference can be used to stop the development of hepatitis in living mice (Cohen 2003). This holds promise for the use of RNAi as a treatment for human disease (Cohen 2003). RNAi could also prove to be useful in cultured cells to better understand cancer by learning about its process of division and signalling (Tuschl 2001).
In addition, the introduction of dsRNA into an organism is easily done. It can be introduced into an organism through either microinjection, electroporation, or even, in the case of the worm Caenorhabditis elegans, by soaking the organism in dsRNA solution (Ullu and Tschudi 2000). It can also be introduced into the worm when it injests bacteria that express dsRNA from recombinant plasmids (Tuschl 2001).
RNAi is especially useful for learning more about organisms that can only be studied with a small number of genetic tools , like the milkweed bug, red flour beetle, planarian, and the freshwater polyp. It will prove useful to examine the evolution of these species (Tuschl 2001).
While RNAi is very useful for understanding the function of a gene, it cannnot replace gene knockout. Though RNAi "mimics" gene knockout in that the target gene is not expressed, it is not like gene knockout because unlike gene knockout in which the gene is deleted, RNAi results in a phenocopy (Waksman Student Scholars 2003). This means that it copies the phenotype as a gene that has no function, but unlike a deletion, the RNAi effects are not inherited in offspring (Waksman Student Scholars 2003). Use of RNAi along with "plasmid transfection technology with inducible vectors" like RNA pol III plasmid systems will make it possible to silence the effects of genes temporarily (Shuey, McCallus, and Giordano 2002).
What are some of the limitations of RNAi ?
RNAi has proven capable of blocking the affects of mRNA in fertilized eggs of Drosophila melanogaster, however, there is some evidence that exists that suggests that RNAi may not work in later stages of mammalian development or in mammalian cells. This is because RNAi may activate the interferon response,"a nonspecific viral defense mechanism," and thus makes RNAi ineffective (Tuschl 2001). Therefore, ways of dealing with the interferon response must be experimented with. These ways may be either using cells lacking the interferon pathway, by finding analogs to dsRNA that do not turn on the interferon response (Tuschl 2001).
References
Ambion®, The RNA Company. RNA Interference and Gene Silencing- History and Overview. May 20, 2002. <http://www.ambion.com/hottopics/RNAi/rnai_may2002_print.html>. Accessed 2003 Feb 10.
Cohen P. 2003 Feb 10. RNAi protects living animals against disease. NewScientist.com<http://www.newscientist.com/news/news.jsp?id=ns99993369>. Accessed 2003 Feb 17.
Cottrell TR, Doering TL. 2003. Silence of the strands: RNA Interference in eukaryotic pathogens. Trends in Microbiology 11:37-43. <http://reviews.bmn.com/rsearch/section/record?uid=TIM.bmn05510_0966842x_v0011i01_02000045&rendertype=text> . Accessed 2003 Feb 12.
Gene Therapy Systems, Inc. Dicer siRNA Generation Kit. <http://www.genetherapysystems.com/catalog/product_line.cfm?product_family_key=1&product_line_key=60>. Accessed 2003 Feb 10.
Montgomery MK. RNAi. <http://www.macalester.edu/~montgomery/RNAi.html#mechanism>. Accessed 2003 Feb 10.
Shuey DJ, McCallus DE, Giordano T. 2002. RNAi: gene-silencing in theraputic intervention. Drug Discovery Today 7: 1040- 1046. <http://reviews.bmn.com/rsearch/section/record?uid=DDT.etd01541_13596446_v0007i20_00002474&node=TOC%40%40DDT%40008%4005%40008_05>. Accessed 2003 Feb 12.
Tuschl T. 2001. RNA interference and small interfering RNAs. Chem Bio Chem 2: 239-145. <http://www.mpibpc.gwdg.de/abteilungen/100/105/RNAi_reviewCBC.pdf>. Accessed 2003 Feb 10.
Ullu E, Tschudi C. 2000. RNAi- applications in parasitology. New technologies for life sciences: A Trends Guide 2000: 43-46. <http://reviews.bmn.com/rsearch/section/record?uid=SUPP.etd00615_14711931_v0001i01_00000073&rendertype=text>. Accessed 2003 Feb 12.
Waksman Student Scholars. <http://avery.rutgers.edu/WSSP/StudentScholars/project/introduction/RNAi.html>. Accessed 2003 Feb 10.
Questions? E-mail Holly Smith at hosmith@davidson.edu