What is a Intein?
Inteins are most commonly described as " protein introns " (www.neb.com 2003). Inteins are sections of post-translational protein that have the ability to excise themselves from the rest of the protein they are incorporated in. Genetically, the DNA coding regions for inteins are within introns, which are within the coding region for the protein in which they are found. However, unlike normal introns, the introns that code for inteins are transcribed into mRNA and translated into a premature protein. A protein is not fully mature until a intein, if incorporated into a protein, is excised from the protein. Inteins can be identified by comparing the DNA sequences of homologous genes against one another. The genes that have intein coding regions within their introns are longer than the homologous genes. Additionally, the coding region in the introns have ten intein motifs that are characteristic to intein encoding introns. (see figure below) The A,N2, B, N4,C,D,E,H,F and G markers are the intein motifs.
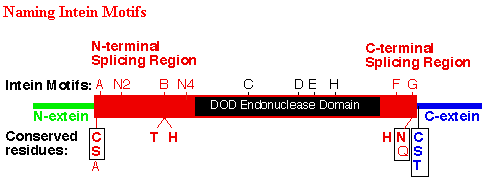
* Permission for use of this figure pending from New England Biolabs http://www.neb.com/ *
How is it that a protein which has a piece of itself excised in from the middle of it's structure function? Before completely disassociating from the protein, the intein fuses the fragments of the protein immediately adjacent to it, exteins, together. The inteins also have the ability to serve as homing endonucleases. The key to understanding the two functions that inteins serve is to first define the structure of the intein and how it can be identified. Inteins are divided into three regions, the N-terminal splicing region, the C-terminal splicing region and the homing endonuclease region (Senejani, et al. 2001). The C-terminal slicing region is bond to the N-terminus of the extein upstream of the intein and the N-terminal region is bound to the C-terminus of the extein downstream of the intein. The endonuclease domain shown above will be explained later. Only the C and N-terminal splicing regions are involved in the excising of the intein from the rest of the protein. (see figure below)
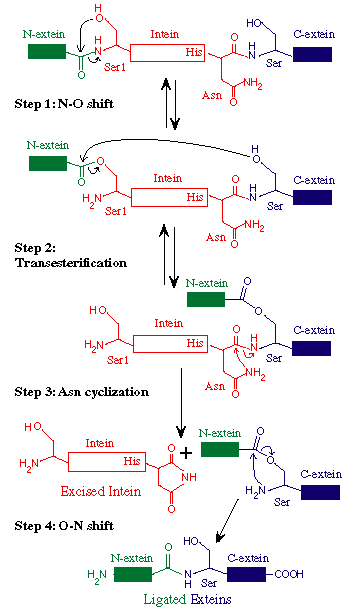
* Permission for use of this figure pending from New England Biolabs http://www.neb.com/ *
The secondary function of the intein is that it serves as a homing endonuclease. This function involves the endonuclease region alone. The process of "homing" is when a endonuclease binds and breaks double stranded DNA at a site on the genome that is specific to that particular endonuclease. After the DNA is broken a segment of DNA removed and is reconstructed. In most cells this is a repair mechanism for the cell to correct any replication errors. In the case of inteins however, the endonuclease region bind to DNA, breaks the double strands and incorporates the intron code which coded for the intein (Gogarten, et al. 2002). The endonuclease only inserts intein coding material at a site in DNA that does not contain intein coding material already. This is why inteins are seen as both transposable elements and genetic parasites. There are also inteins that are much smaller than regular inteins, only 130-200 aa long, and do not contain any endonuclease region. These are called "mini-inteins" and they only possess the ability to excise themselves from the protein they were incorporated into and rebind the exteins that are adjacent to it.
Inteins are only found in microorganisms which include mycobacterium, thermophilic archaebacteria, baker's yeast, and chloroplasts in red algae. All of these organisms are not only microscopic but they are also ancient organisms. When analyzed the Guanine-Cytosine content found in the intein coding region is quite divergent from the extein coding region (www.neb.com 2003) that inteins are adjacent to, but intein are very similar to other intein coding regions in other species which implies horizontal transfer of genetic material from one species of organism to another.(Gogarten, et al. 2002)
What are the potential uses of Inteins?
The use of inteins in experimental biology has been primarily been to extract purified proteins from prokaryotes and to construct proteins that are cyclized. Cyclized protein according to polymer theory (Flory, 1956) have increases stability over its linear state. The company New England BioLabs created a system that utilizes the inteins to purify target proteins in a column purification without the use of proteases which may damage target proteins. (see figure below)
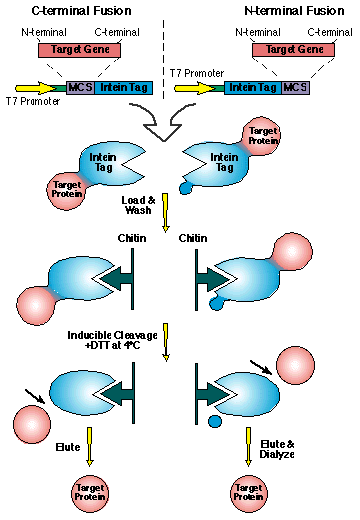
* Permission for use of this figure pending from New England Biolabs http://www.neb.com/ *
The target gene that is to be translated into a protein and purified is linked to either the C or N-terminal splicing region of the intein sequence. In addition a T7 promoter is created upstream of both the target protein and intein region. The T7 promoter controls the expression level of the gene construct. The intein tag has a chitin binding domain in it which allow it to bind to chitin which are present in a purification column made by New England Biolabs. The protein splicing properties of the inteins are then induced by either DTT, change in pH or a change in temperature within the column. The target protein then dissassociates from the chitin bound intein and the target protein precipitates through the column into a catch tube.
The protein splicing properties of inteins are also manipulated to cyclize proteins by creating inteins on ether side a target protein which links the C and N-terminus of a target protein together to fortify the stability of the protein. (see figure below) The figure below is the system that is produced by New England Biolabs which uses their chitin binding technology. Notice dynamics of the system and how the construct is able to isolate a target protein and expose its naked C and N-termini.
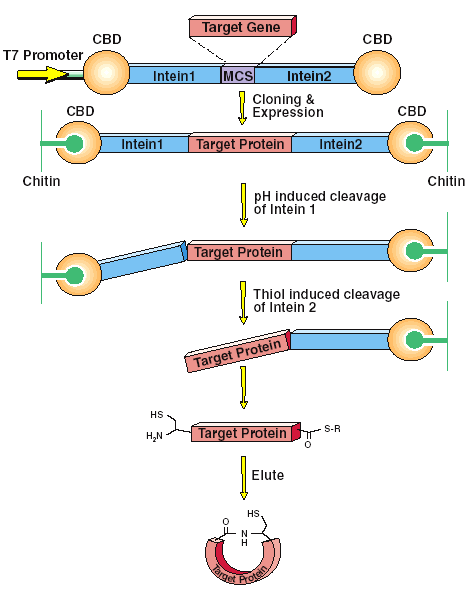
* Permission for use of this figure pending from New England Biolabs http://www.neb.com/ *
Links