This web page was produced as an assignment for an undergraduate course at Davidson College.
ATP Synthase Orthologs
What is an Ortholog?
"Orthologs are genes in different species that evolved from a common ancestral gene by speciation. Normally, orthologs retain the same function in the course of evolution. Identification of orthologs is critical for reliable prediction of gene function in newly sequenced genomes" (Christopher Lewis).
Some Information on ATP Synthase Orthologs:
There are four classes of ATPases. Different types of ATPases differ in structure and function depending where they are found. V-type ATPases have a peripheral domain (V1) and an integral membrane component (Vo). V1 has seven subunits. Vo also has three types of subunits. This is very similar in structure to F-Type ATPases (like ATP Synthase), but V-type ATPases are located in vacular membranes of higher plants and funghi and in lysosomal, endosomal, and secretory vesicle membranes of animals, rather than in the mitochondrial inner membrane. ATP Synthase is found in the thylakoid membrane of higher plants, in the plasma membrane of prokaryotes, and in the mitochondrial membrane of eukaryotes. ATP Synthase always functions to catalyze the formation of ATP from ADP and Pi. (Nelson et al., 2000)
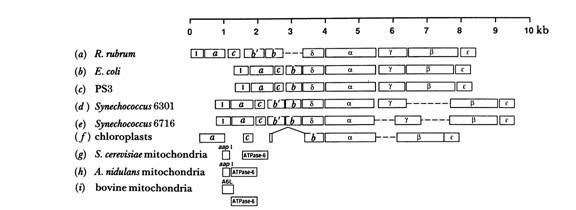
Fig. 1. Organization of genes for subunits of ATP Synthase in mitochondria, chloroplasts, and eubacteria. (Walker, et al. 1990)
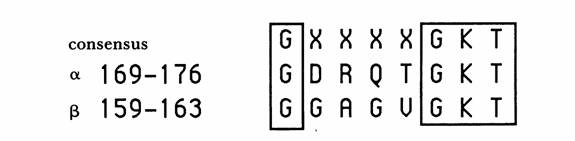
Fig. 2. The purine nucleotide binding sequence motif in the alpha and beta subunits of ATP Synthase. The numbers correspond with the E. Coli enzyme but the residues were conserved in all species investigated by Walker, et al. (Walker, et al 1990)
Table 1. Membrane Domain Of The Subunit B Of The E. Coli ATP Synthase. Click PDB for the full nucleotide sequence. Data courtesey of the Protein Data Bank. Using BLAST, this sequence popped up in many other bacteria including Salmonella typhimurium, Erwinia carotovora, and Yersinia pseudotuberculosis.
| Chains | Residues | Mol. Weight [D] | Chain Type | DBREF |
|---|---|---|---|---|
| 1B9U:A | 34 | 3683 | Protein | SwissProt: ATPF_ECOLI |
Table 2. Bovine Mitochondrial F1 ATPase. Click on PDB the for the full nucleotide sequences. Data courtesey of the Protein Data Bank.
| Chains | Residues | Mol. Weight [D] | Chain Type |
|---|---|---|---|
| 1BMF:A | 510 | 55222 | Protein |
| 1BMF:B | 510 | 55222 | Protein |
| 1BMF:C | 510 | 55222 | Protein |
| 1BMF:D | 482 | 51693 | Protein |
| 1BMF:E | 482 | 51693 | Protein |
| 1BMF:F | 482 | 51693 | Protein |
| 1BMF:G | 272 | 30126 | Protein |
Table 3. Rat Liver F1-ATPase. Click on PDB for the full nucleotide sequences. Notice the difference in residues and molecular weight when compared to the mitochondrial ATPase in cows. Data courtesey of the Protein Data Bank.
| Chains | Residues | Mol. Weight [D] | Chain Type |
|---|---|---|---|
| 1MAB:A | 390 | 42169 | Protein |
| 1MAB:A:1 | 510 | 55270 | Protein |
| 1MAB:B | 479 | 51341 | Protein |
| 1MAB:G | 270 | 29919 | Protein |
Table 4. Structure Of The Chloroplast F1-ATPase From Spinach. ATP Synthase in plants is used mostly in photosynthesis, especially if it is located in the chloroplast. It serves the same function of producing ATP, but the process and environment are very different from bovine mitochondria or rat liver. The sequence should not be too different since the sequence determines the form and form fits function and the function does not change. Click on PDB for the full nucleotide sequences. Data courtesey of the Protein Data Bank.
| Chains | Residues | Mol. Weight [D] | Chain Type | DBREF |
|---|---|---|---|---|
| 1FX0:A | 507 | 55440 | Protein | SwissProt: ATPA_SPIOL |
| 1FX0:B | 498 | 53856 | Protein | GenBank: AAA84626 |
Table 5. Structure Of The Regulatory Subunit H Of The V-Type ATPase Of Saccharomyces Cerevisiae. Click on PDB for the full nucleotide sequence. Compare to F-type ATPase. As stated above, the V-type ATPase is not located in the mitochondrial inner membrane but rather in vacular and secretory vesicle membranes. Like in the spinach, this changes the environment and process, but does not change the fact that ATP Synthase produces ATP from ADP. Data courtesey of the Protein Data Bank.
| Chains | Residues | Mol. Weight [D] | Chain Type | DBREF |
|---|---|---|---|---|
| 1HO8:A | 480 | 54548 | Protein | SwissProt: VATH_YEAST |
ATP Synthase Mutations and their Effects:
ATP Synthase is vital to every organism. Glycolysis alone cannot produce enough ATP to sustain life. So, any mutation that causes a loss of or change in function of ATP Synthase will generally be fatal. Two antibiotics, Oligomycin and venturicidin, are known to bind ATP Synthase and consequently inhibit ATP synthesis. They are toxic. (Nelson et al., 2000)
Originally it was thought that all ATPase defects were caused by inherited mtDNA mutations in the F0 complex subunit 6. But now it is known that biosynthesis can cause a selective defect of mitochondrial ATP Synthase that causes fatal neonatal lacticacidemia with cardiomegaly and helptomegaly. (McKusick, 1999)
NARP syndrome may be caused by a point mutation at nucleotide 8993 on the gene encoding subunit 6 of mitochondrial ATP Synthase. A correlation between clinical severity and the amount of mutant mitochondrial DNA in patients has been observed.(Kelly, 2000).
For background information on ATP Synthase's structure and function, please refer to my "My Favorite Protein" page and the references listed on it.
References:
Kelly, J. Narp Syndrome. 2000. Accessed on OMIM database,(#551500). 2005 March 8.
McKusick, VA. ATP Synthase Deficiency. 1999. Accessed on OMIM database, (#604273). 2005 March 7.
NCBI. 2005. <http://www.ncbi.nlm.nih.gov/> Accessed 8 March 2005.
Nelson, DL, Cox MM. Lehninger Principles of Biochemistry, third edition. New York: Worth Publishers, 2000. 675.
Protein Data Base. < http://www.rcsb.org/pdb/> Accessed 8 March 2005.
Walker, J.E, et al. (1990) Structural Aspects of Proton Pumping ATPases. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences: 326, 367-378.
© Copyright 2002 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: jecarlson@davidson.edu