This web page was produced as an assignment for an undergraduate course
at Davidson College.
Reverse Transcriptase
"Is function the mechanical result of form, or is form merely the manifestation of function or activity? What is the essence of life--organization or activity?"
- Étienne Geoffroy St. Hilaire (1772-1844)
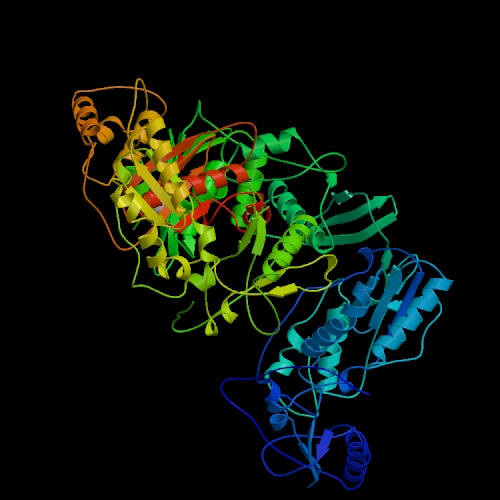
Figure 1. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Image courtesy of Protein Databank. ID: 1hmv; PDB, 2005.
DNA Polymerases--An Overview:
DNA polymerases are enzymes that make a complimentary DNA strand from a DNA/RNA template, adding DNA nucleotides to the 3'-end of the appropriate primer. While there are several different specific DNA polymerases, many can be grouped into four families based on homologous sequences: pol I, pol a, polymerase family X, and reverse transcriptase. These four families all share some structural similarities, including a U-shaped DNA binding cleft that has "fingers," "thumb," and "palm" subdomains (Doublie, Sawaya, & Ellenberger, 1999). Doublie, Sawaya, & Ellenberger (1999) compared the structures of four different polymerases from three different families (See Figure 2.) and found that although the amino acid sequences of these enzymes were very dissimilar, they all had metal ions (usually magnesium) in their active site. During nucleotidyl transfer, these metal ions play a key role in interacting with the phosphates of the nucleotides and the 3'-end of the primer. These metal binding sites are highly conserved among different DNA polymerases, which highlights their importance for proper function of nucleotide polymerization.
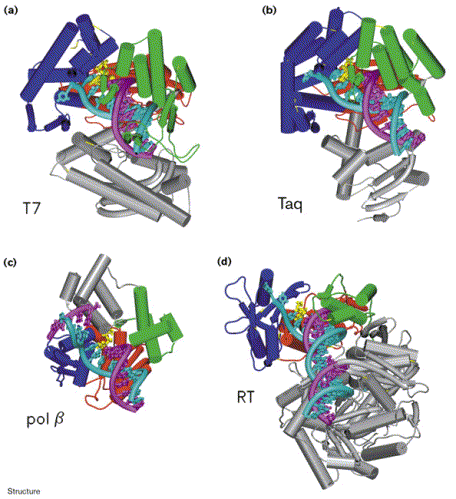
Figure 2. Comparison of the structures of four different DNA polymerases in complex with DNA: (a) T7 DNA polymerase, (b) Taq polymerase, (c) pol b, and (d) HIV-1 Reverse Transcriptase (p66 subunit only). DNA is shown in cyan and magenta, and the nucleotide substrates in yellow. The fingers subdomain is shown in dark blue, the thumb in green, and the palm in red. Image courtesy of Structure.
Reverse Transcriptase:
Unlike other DNA polymerases, reverse transcriptase (RT) is unique in that it is able to make a complimentary DNA strand from an RNA template. Reverse transciptases are usually found in retroviruses, which use RT to make a DNA copy of their own RNA genome, which is then incorporated into the host cell's genome. The most notorious of these retroviruses is HIV-1, which causes AIDS (Hartwell et al., 2004).
Structure:
Reverse transcriptase is a heterodimer protein (composed of 2 different polypeptides) that has a 66 kDa subunit (p66) and a 51 kDa subunit (p51) (Hsiou et al., 1996). Subunit p66 has an N-terminal polymerase domain (440 residues) and a C-terminal RNase H domain (120 residues) (See Complete Amino Acid Sequence). It also contains the DNA-binding cleft. Both subunits have a polymerase domain, but it is only functionally active in p66. Subunit p51 is simply a cleaved version of p66, and lacks the RNase H domain. Furthermore, subunits p66 and p51 have identical amino acid sequences (p51 shortened), but are structurally very different (See Figure 3) (Goldman & Marcey, 2001). Both subunits p66 and p51 each have 4 subdomains (fingers, thumb, palm, and connection) which are arranged differently in the two subunits (See Figure 4) (Hsiou et al., 1996). The connection and palm subdomains each contain 3 beta sheets with alpha helices. The thumb subdomains contain 3 alpha helices (Goldman & Marcey, 2001). The polymerase active site of subunit p66 is highlighted by 3 catalytic residues (Asp110, Asp185, Asp186) that may play a role in binding metal ions, and hence nucleotidyl phosphate interactions (Hsiou et al., 1996; Goldman & Marcey, 2001).
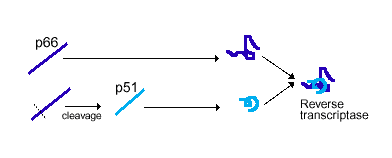
Figure 3. Subunits p66 and p51 have identical amino acid sequences, but p51 is cleaved to produce a shorter polypeptide strand. The two subunits then structurally conform differently before they combine to produce the final product, reverse transcriptase. Image courtesy of Dan Stowell .
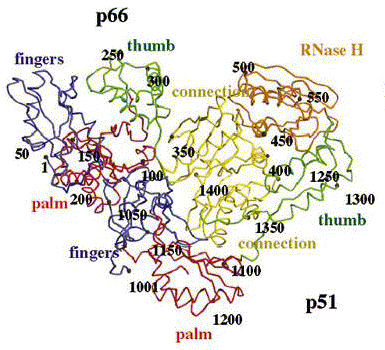
Figure 4. Reverse transcriptase showing both the p66 and p51 subunits. Both subunits have the palm, thumb, fingers, and connection subdomains. The RNase H domain is located on the p66 subunit only. Every 50th amino acid residue is labeled. Image courtesy of Structure.
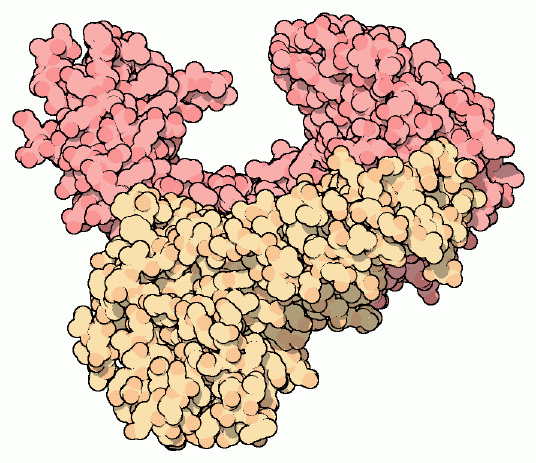
Figure 5. Drawing of unliganded reverse transcriptase. Subunit p66 is shown in dark pink, and subunit p51 is shown in creme. Notice the similarity of structure with Figure 4. Image courtesy of David S. Goodsell.
Function:
The primary function of reverse transcriptase is to build a DNA strand from an RNA template, performed at the polymerase active site of subunit p66. Most of the nucleotidyl interactions that occur in reverse transcriptase occur between the RNA template and subunit p66. The thumb and fingers subdomains of the p66 subunit serve as a clamp to hold RNA in the polymerase active site of the palm subdomain (See Figure 6) (Goldman & Marcey, 2001). The trio of aspartic acid residues (110, 185, 186) at the polymerase active site ligate two metal ions (possibly magnesium), which interact with the phosphates of the DNA primer and incorporated nucleotides (Doublie, Sawaya, & Ellenberger, 1999). The p51 subunit serves as a binding site for the anti-codon stem and loop of tRNAlys which functions as the primer to begin DNA synthesis (Goldman & Marcey, 2001). Once the cDNA has been built from the RNA template, the RNA is cleaved into pieces at the RNase H nuclease site (See Figure 6). Once the RNA is cleaved from the complex, a second DNA strand is made, complimentary to the first DNA strand, to form the final double helix DNA molecule. This process again occurs at the polymerase active site on subunit p66 (Goodsell, 2002).
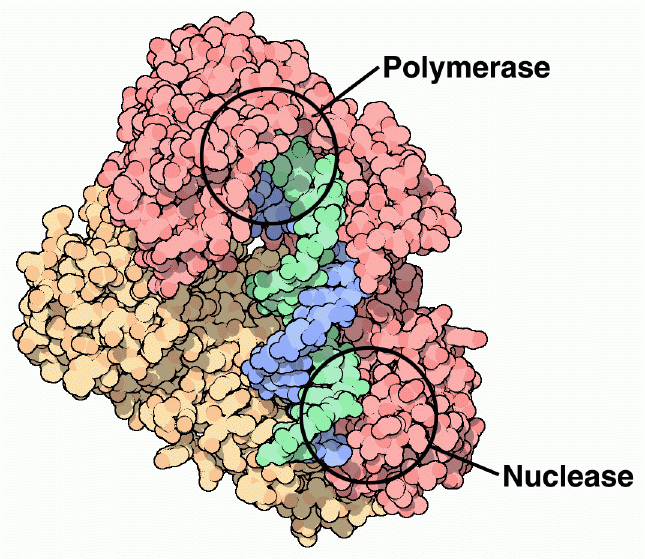
Figure 6. Reverse transcriptase has a polymerase active site that polymerizes new DNA strands from an RNA template, and a nuclease site, which cleaves the RNA template into pieces. Subunit p66 is shown in dark pink, and subunit p51 is shown in creme. The nucleic acid substrate is shown in green and blue. Image courtesy of David S. Goodsell.
Human Immunodeficiency Virus-1:
As a retrovirus, HIV-1 uses reverse transcriptase (See Figure 7) to copy its own RNA genome and make complimentary DNA, which it then incorporates into the host cell's genome. Because RT plays such a pivotal role in HIV-1 activity, it is often a prime target for viral inhibition. There are generally two categories of RT inhibitors. The first category of inhibitors are nucleoside analogs (ddNTPs), which once added to the DNA strand, prevent RT from polymerizing the rest of the DNA (See Figure 8 ). The second category of inhibitors are non-nucleoside analog inhibitors, which play a role in interacting with a specific site near the polymerase active site of p66 (See Figure 9) (Ren et al., 1995). The problem with nucleoside analogs is that they are not HIV-1 RT specific, and therefore can interfere with other DNA polymerases, which can cause serious side effects. Non-nucleoside analog inhibitors are much more specific for inhibiting HIV-1 RT only, and have much fewer side effects. Still, neither of the two categories of inhibitors are complete cures for HIV-1, as the virus is often able to produce drug-resistant mutants (Ding et al., 1995).
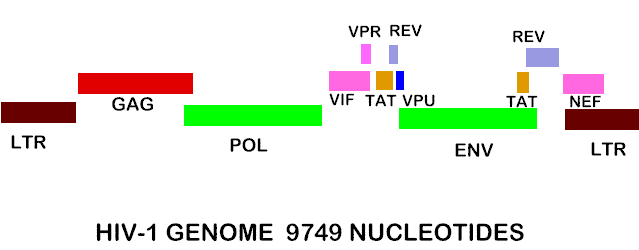
Figure 7. Shows the complete RNA genome of HIV-1. The POL gene codes for reverse transcriptase. You can also view the HIV-1 genome at NCBI Entrez Genome. Image courtesy of Dr. Richard Hunt.
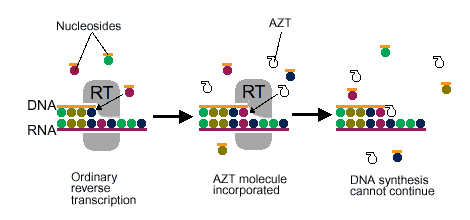
Figure 8. Shows a nucleoside analog inhibitor (AZT) preventing reverse transcriptase from continuing the synthesis of DNA. Image courtesy of Dan Stowell .
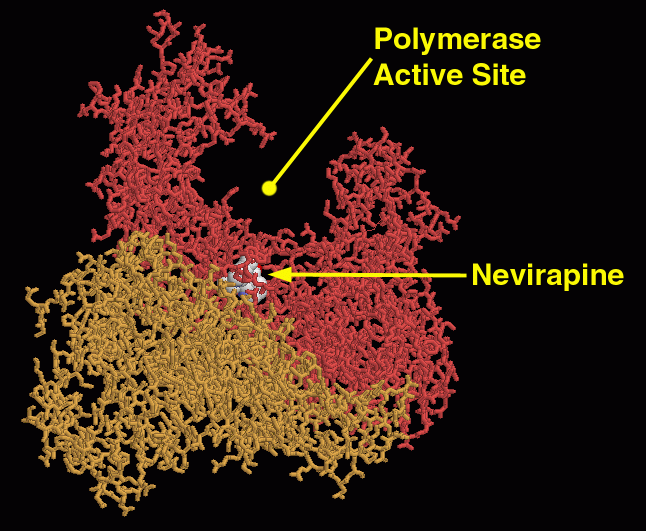
Figure 9. A non-nucleoside analog inhibitor (Nevirapine) interacts with a specific site near the polymerase active site to interfere with reverse transcriptase function. Image courtesy of David S. Goodsell.
One of the reasons why HIV-1 is so hard to inhibit and often resistant to drugs is because of its reverse transcriptase. Unlike many other DNA polymerases, RT is very inaccurate when it polymerizes new DNA strands. It usually makes 1 mistake about every 5000 nucleotides it incorporates into a DNA strand, compared with DNA polymerase which makes 1 mistake about every million nucleotides. Such inaccuracy by RT leads to mutations in the HIV-1 genome which are advantageous for the virus to continually build resistance to the host immune system and new drug inhibitors (Hartwell et al., 2004).
Ding J, Das K, Tantillo C, Zhang W, Clark, Jr AD, Jessen S,
Lu X, Hsiou Y, Jacobo-Molina A, Andries K, Pauwels R, Moereels H, Koymans
L, Janssen PAJ, Smith, Jr RH, Groeger Koepke M, Michejda CJ, Hughes SH, Arnold
E. (1995). Structure of HIV-1 reverse transcriptase in a complex with non-nucleoside
inhibitor a-APA R 95845 at 2.8 A resolution. Structure 3(4). <http://www.sciencedirect.com/science?_ob=ArticleURL&_aset=B-WA-A-W-AB-MsSAYWA-UUA-AAAAUVBBVD-AAUYZWVAVD-EWAVDBCAC-AB-U&_rdoc=11&_fmt=full&_udi=B6VSR-4CXD831-19&_coverDate=04%2F30%2F1995&_cdi=6269&_orig=search&_st=13&_sort=d&view=c&_acct=C000058476&
_version=1&_urlVersion=0&_userid=2665120&md5=5fc2086515e06e1e192a74bf524e9bde>
Accessed 2005 Feb 12.
Doublie S, Sawaya MR, Ellenberger T. 1999. An open and closed
case for all polymerases. Structure (7)2. <http://www.sciencedirect.com/science?_ob=ArticleURL&_aset=B-WA-A-W-AB-MsSAYWA-UUA-AAAAUVBBVD-AAUYZWVAVD-EWAVDBCAC-AB-U&_rdoc=4&_fmt=full&_udi=B6VSR-4CXCWVD-T&_coverDate=02%2F15%2F1999&_cdi=6269&_orig=search&_st=13&_sort=d&view=c&_acct=C000058476&_
version=1&_urlVersion=0&_userid=2665120&md5=260675e1d2914c88e9789eb57c0e2e4f>.
Accessed 2005 Feb 12.
Goldman M, Marcey D. 2001. HIV-1 Reverse Transcriptase. The Online Molecular Museum. <http://www.clunet.edu/BioDev/omm/hivrt/hivrt.htm>. Accessed 2005 Feb 12.
Goodsell, DS. 2002. Molecule of the month: reverse transcriptase. RCSB Protein Data Bank. <http://www.rcsb.org/pdb/molecules/pdb33_1.html>. Accessed 2005 Feb 12.
Hartwell LH, Hood L, Goldberg ML, Reynolds AE, Silver LM, Veres RC. Genetics: from Genes to Genomes, 2nd Edition. Boston: McGraw Hill, 2004. pp. 252-253.
Hsiou Y, Ding J, Das K, Clark, Jr AD, Hughes SH, Arnold E. 1996.
Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications
of conformational changes for polymeraztion and inhibition mechanisms. Structure
4(7). <http://www.sciencedirect.com/science?_ob=ArticleURL&_aset=B-WA-A-W-AB-MsSAYZW-UUA-AAAAUWYBBE-AAUYZUEABE-EWAWBCEEY-AB-U&_rdoc=7&_fmt=full&_udi=B6VSR-4CJB814-1M&_coverDate=07%2F31%2F1996&_cdi=6269&_orig=search&_st=13&_sort=d&view=c&_acct=C000058476&_
version=1&_urlVersion=0&_userid=2665120&md5=563c490ff542b90bcd5e48547debf041>
Accessed 2005 Feb 12.
Ren J, Esnouf R, Hopkins A, Ross C, Jones Y, Stammers D, Stuart
D. 1995. The structure of HIV-1 reverse transcriptase complexed with 9-chloro-TIBO:
lessons for inhibitor design. Structure 3(9). <http://www.sciencedirect.com/science?_ob=ArticleURL&_aset=B-WA-A-W-AB-MsSAYWA-UUA-AAAAUVBBVD-AAUYZWVAVD-EWAVDBCAC-AB-U&_rdoc=10&_fmt=full&_udi=B6VSR-4CXD8FR-3D&_coverDate=09%2F30%2F1995&_cdi=6269&_orig=search&_st=13&_sort=d&view=c&_acct=C000058476&_
version=1&_urlVersion=0&_userid=2665120&md5=5141061af964eeec50a695106ea0e7ba>.
Accessed 2005 Feb 12.
Davidson College Molecular Biology Homepage
My Molecular Biology Home Page
© Copyright 2005 Department of Biology, Davidson College,
Davidson, NC 28035
Send comments, questions, and suggestions to: kekoike@davidson.edu;
macampbell@davidson.edu
