*This website was produced as an assignment for an undergraduate course at Davidson College.* |
BRCA1 Orthologs
What is an Ortholog?
Orthologs are genes in two or more species that evolved from a common ancestral gene by speciation. Normally, orthologs retain the same function in the course of evolution. Identification of orthologs is critical for reliable prediction of gene function in newly sequenced genomes (Lewis, 2005 ). According to recent research, hBRCA1 is involved in 20% - 45% of inherited breast cancer cases and it is implicated in many mechanisms involved in response to DNA damage (Lafarge et. al., 2003). To date, in order to have a better understanding of the structure-function of this protein, BRCA1 orthologs have been characterized in other genomes.
BRCA1 orthologs have been defined by the presence of one N-terminal RING and two C-terminal BRCT domains (Orelli et. al., 2001). Therefore In order to search for putative orthologs the typical hBRCA1 structural organization of these three domains has to be considered.

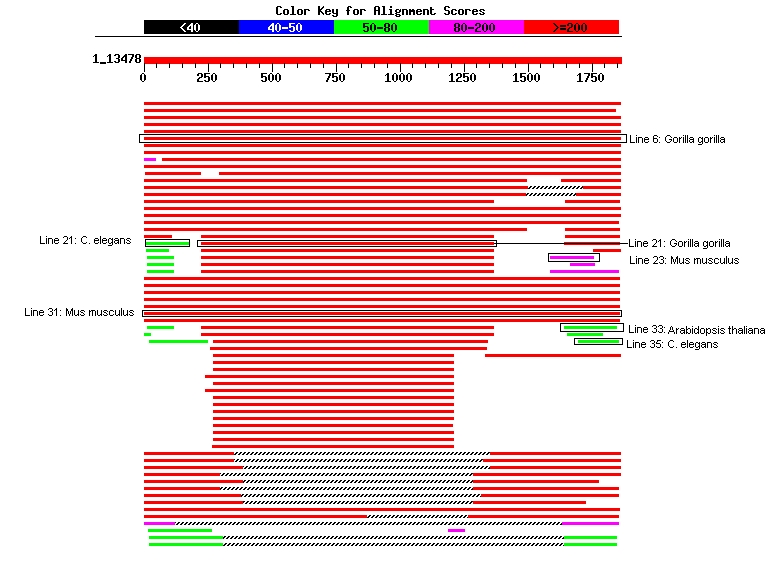
Fig. 1 An overview of different database sequences aligned to the BRCA1 sequence. The score of each alignment is indicated by one of five different colors, which divides the range of scores into five groups. Multiple alignments on the same database sequence are connected by a striped line (Source: Image adapted from a courtesy of NCBI) note:due to the nature of the website, permanent link not available The known functions of hBRCA1 are nearly all linked to RING and BRCT domains: with an ubiquitin E3 ligase activity and interactions with several proteins in the RING domain, while the BRCT domains are involved in transcriptional transactivation and are also required for interaction with several proteins. This BRCT domain is a signature of proteins involved in DNA repair and is present in numerous species, including vertebrates, yeast and plants, suggesting a conserved role for this domain. As shown by Joukov et al., the Xenopus structural ortholog of BRCA1 was also reported as a functional ortholog. So, conserved domains strongly indicated that this structural organization is the signature of BRCA1 orthologs, and it is linked to conserved activities and function.
Examples of Mammalian Orthologs:
Gorilla Gorilla
- AAT44835 breast cancer type1, E=0.0 (see Fig.1,Line 6)
- AAC39584 BRCA1, E= 0.0 (see Fig. 1, Line 21)
Mus Musculus
- AAH03248 Unknown, E= 3e-46 (see Fig. 1, Line 23)
- AAC52323 breast cancer 1, E= 0.0 (see Fig.1, Line 26)
- AAD00168 Brca1, E= 0.0 (see Fig 1, Line 27)
- AAH68303 Breast Cancer 1, E= 0.0 (see Fig.1, Line 31)
Orthologs of hBRCA1 had been described only in vertebrates: in mammalian (Szabo et. al., 1996), Xenopus (Joukov et. al., 2001) and Gallus genomes (Orelli et. al., 2001). The orthologs of BRCA1 in Arabidopsis thaliana and in Caenorhabditis elegans have also been identified, showing a highly similar structure to hBRCA and the same characteristic molecular organization (see table below).
The ortholog of BRCA1 in C. elegans encodes a more divergent protein that is significantly shorter than BRCA1 but is most related to its exon delta 11 splice variant. The discovery of functional orthologs of the BRCA1 tumor suppressor gene and its partner BARD1 in C. elegans suggest that Ce-brc-1 and Ce-brd-1 function together in a common DNA repair pathway, consistent with the role of BRCA1 and BARD1 in mammalian cells (Boulton et. al., 2004).
Examples of Orthologs in Other species:
Arabidopsis Thaliana (plant)
- D86171 hypothetical protein (Imported), E= 1e-11 (see Fig.1, Line 33)
Sequence alignment AtBRCA1 confirmed the presence of one RING domain between amino acids 16 and 53 and two BRCT domains between amino acids 727 and 809, and 842 and 935. Alignment of AtBRCA1 and hBRCA1 showed 34% identity–61% similarity in the RING domain, and 28% identity–61% similarity in the BRCT region. To date, AtBRCA1 is the first non-vertebrate BRCA1 ortholog (Doutriaux et. al., 1998).
Caenorhabditis elegans (nematode)
- T19770 hypothetical protein C36A4.8, E= 9e-09
(see Fig. 1, Line 21)
Sequence alignment Ce-BRC-1 is a protein of 596 amino acids that is 24% identical and 52% similar to the major human BRCA1 delta-exon 11 splice variant. Similar to BRCA1, Ce-BRC-1 contains an N-terminal RING finger, a nuclear localization signal, and two C-terminal BRCT repeat domains (Boulton et. al., 2004).- AAO51148 similar to Y55B1BR.3.p E= 2e-05 (see Fig. 1, Line 21)
BRCA1 orthologs, between human and other species, presented slight differences in length and/or in structural composition, but domains linked to protein activities are always conserved within the same structural organization. Analysis of other orthologs showed that sequence conservation is particularly important in the RING and BRCT domains but less significant between them. This may suggest that the BRCA1 central region is species-specific (Orelli et. al., 2001).
In summary, the existence of BRCA1 orthologs, suggests that identification of the BRCA1 pathway in will lead to important insights into its role in the DNA repair processes in human disease.
References:
- Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, and Vidal M. 2004 BRCA1/BARD1 Orthologs Required for DNA Repair in Caenorhabditis elegans. Current Biology 14: 33-39. doi:10.1016/j.cub.2003.11.029
- Doutriaux MP, Couteau F, Bergounioux C, and White C. 1998 Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257 : 283–291.
- Joukov V, Chen J, Fox EA, Green JB, and Livingston DM. 2001 Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl Acad. Sci. USA 98: 12078–12083. < http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=59770 >
- Lafarge S, and Montané MH. 2003 Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Research 31(4): 1148–1155 <http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=150221>
- Lewis C. Feb 2005. Homepage. < http://homepage.usask.ca/~ctl271/857/def_homolog.shtml > Accessed 2005, March 3.
- Orelli BJ, Logsdon JM, and Bishop DK. 2001 Nine novel conserved motifs in BRCA1 identified by the chicken orthologue. Oncogene 20: 4433–4438.
- Szabo CI, Wagner, LA, Francisco LV, Roach JC, Argonza R, King MC, and Ostrander EA. 1996 Human, canine and murine BRCA1 genes: sequence comparison among species. Hum. Mol. Genet. 5: 1289–1298. <http://hmg.oupjournals.org/cgi/content/full/5/9/1289>
Visit Davidson College Biology Department Home Page
Visit Molecular Biology Home Page
Send comments, questions, and suggestions to: Daniela V. Alvarez