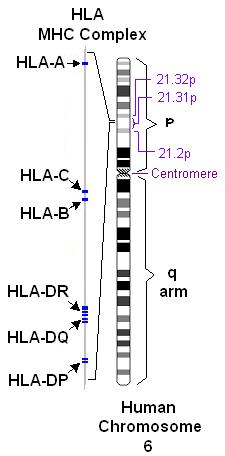
If you read my previous webpage describing MHC class I, you know that there are hundreds of alleles that can code for the α subunit of MHC class I in humans. While the general structure of all MHC class I molecules remains similar from one allele to another, the amino acid sequences of the α subunit must differ among alleles to generate MHC's ability to bind to such a wide range of antigens. Remember that MHC molecules in humans are called "human leukocyte antigen" (HLA) system. An HLA gene was first sequenced and characterized by Malissen et al., 1982, and now we know that one human can possess up to six different MHC class I α subunit alleles - three from each parent. These alleles are located on chromosome 6 and are designated as HLA-A, HLA-B, and HLA-C (Lamm et al., 1974; Figure 1). Despite the huge number of HLA alleles in a population, these proteins generally only differ in amino acid sequence at the antigen binding groove. Thus, it is conceivable that MHC class I genes would be conserved in closely related species. This webpage examines the orthology† of MHC class I using an HLA-A sequence as a reference.

Figure 1. HLA alleles on human chromosome 6. HLA-A, B, and C represent the three MHC class I α subunit alleles, while HLA-DR, DQ, and DP are MHC class II alleles. This image was obtained from Wikimedia Commons.
The evolution of HLA genes has been well-characterized. Though the exceptional diversity of MHC genes stems from an incredibly high rate of mutation as well as gene duplication, it is evident that many HLA polymorphisms arose long before the divergence of humans and apes (Klein and Figueroa, 1986; Lawlor et al., 1988). In fact, it has been shown that an HLA allele is more likely to be related to a chimpanzee MHC class I allele than another human allele for the same HLA gene. This evidence implies that many MHC genes appeared early in the evolution of vertebrates, so a high degree of orthology among some MHC class I genes is probable, though the number of genes and allelic diversity will vary among species due to the high level of mutation of MHC class I and the tendency for gene duplication. To assess the level of orthology of a particular HLA gene, I obtained the amino acid sequence of an HLA-A gene from NCBI, and I performed a BLASTp to verify the gene as a Homo sapiens HLA-A (Figure 2).
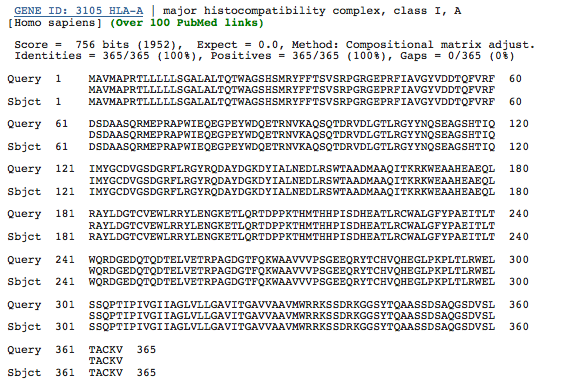
Figure 2. BLASTp of an HLA-A amino acid sequence obtained from NCBI. The sequences in the databases are identical, verifying that this is the sequence of an HLA-A gene in humans.
 A |
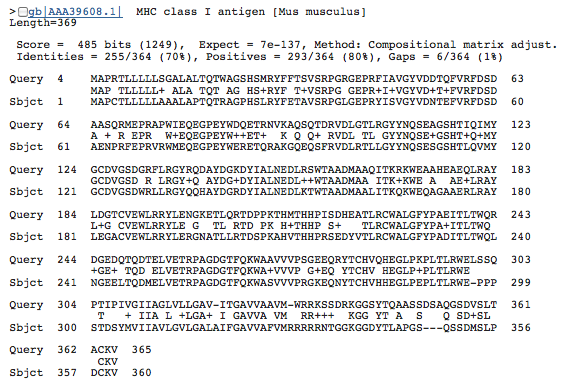 B |
 C |
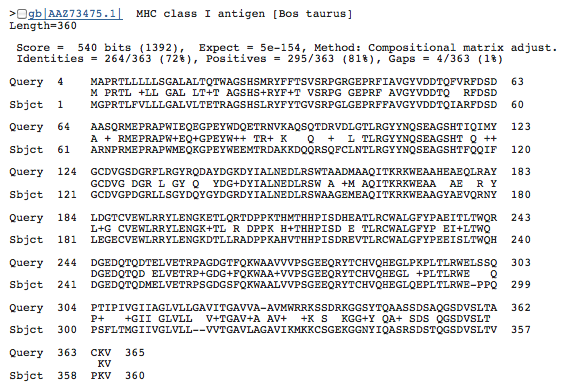 D |
Figure 3. BLASTp of an HLA-A amino acid sequence found this gene in many other species: common chimpanzee (A), mouse (B), canine (C), and cow (D). Each of these BLASTp results are similar in amino acid length (approximately 350-365 aa) and have an extremely low e value, suggesting that these genes are orthologous.
Kasahara M, M Hayashi, K Tanaka, H Inoko, K Sugaya, T Ikemura, and T Ishibashi. Chromosomal localization of the proteasome Z subunit gene reveals an ancient chromosomal duplication involving the major histocompatibility complex.Proc. Natl. Acad. Sci. USA 93: 9096-9101.
Klein J and F Figueroa. 1986. Evolution of the major histocompatibility complex. CRC Crit. Rev. Immun. 6: 295-386.
Lamm LU, U Friedrich, BG Petersen, J Jorgensen, J Nielsen, AJ Therkelsen, and F Kissmeyer-Nielsen. 1974. Assignment of the major histocompatibility complex to chromosome no. 6 in a family with a pericentric inversion. Hum. Hered. 24: 273-284.
Lawlor DA, FE Ward, PD Ennis, AP Jackson, P Parham. 1988. HLA-A and B polymorphisms predate the divergence of humans and chimpanzees. Nature 335: 268-271.
Malissen M, B Malissen, and BR Jordan. 1982. Exon/intron organization and complete nucleotide sequence of an HLA gene. Proc. Natl. Acad. Sci. 79: 893-897.
†Orthology refers to the presence of a gene in different species; this gene derives from a common ancestor.