Orthologs of the Human Insulin-Like Growth Factor 1 Gene
Goal: To understand what an ortholog is, and to examine orthologs of the Human IGF-1 Gene
Defining an ortholog is no simple task. Let's take a look at some of the recent literature to attempt
to understand where biology's definition of an ortholog now lies.
Orthologs can be defined as "genes that have diverged after a
speciation event... [that] tend to have similar function" (Fulton et al. 2006).
Thus, orthologs are genes whose encoded proteins fulfill similar roles
in different species. The importance of orthologs is quite simply seen
when imagining a hypothetical comparison of two genes, A and B, that
encode proteins with similar functions in two different species (human
and chimp, for example). If one compares the two protein sequences
encoded by the orthologs, the truly critical parts of the gene will be
conserved. What has remained constant can probably be interpreted as
crucial to the functioning; what has changed, minor.
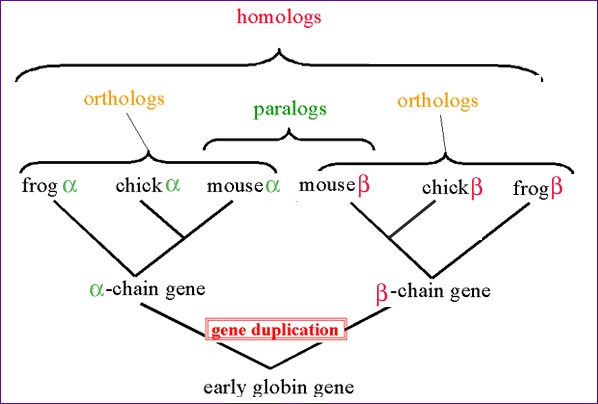
Figure 1. A visual representation of orthologs (and some other commonly confused terms, paralogs and homologs). Permission pending from UW-Lacrosse Biology Department.

Figure 1. A visual representation of orthologs (and some other commonly confused terms, paralogs and homologs). Permission pending from UW-Lacrosse Biology Department.
With the definition of an ortholog now
clear and an understanding of their potential usefulness, we look to apply our understanding of orthologs to an
interesting protein examined earlier - Human Insulin-Like Growth Factor 1.
This (clearly) being a human gene, a search of the seven species of
particular interests to geneticists and biologists should be
particularly interesting.
In humans, IGF-1 is a 70 amino acid long protein. This is quite short. Thus, the regions of similarity (and importance) should be relatively simple to identify. Researchers have identified orthologs of the IGF-1 gene in numerous species, including mice, zebrafish, rats, cows, and chimps ("Insulin-like growth factor 1 (somatomedin C)").
A Blast query using the protein sequence of IGF-1 reveals some interesting things. Among the big seven genomic species, only two - Drosophila melanogaster (the fruit fly) and Mus musculus (the mouse) - produced any sort of meaningful sequence alignments. There appear to be no orthologs of IGF-1 in prokaryotes ("Insulin-like growth factor 1 (somatomedin C)").
Davidson College
Davidson Molecular Biology 2010 Home
Michael Rydberg's Molecular Home
Please contact me with any comments or questions at mirydberg@davidson.edu
In humans, IGF-1 is a 70 amino acid long protein. This is quite short. Thus, the regions of similarity (and importance) should be relatively simple to identify. Researchers have identified orthologs of the IGF-1 gene in numerous species, including mice, zebrafish, rats, cows, and chimps ("Insulin-like growth factor 1 (somatomedin C)").
A Blast query using the protein sequence of IGF-1 reveals some interesting things. Among the big seven genomic species, only two - Drosophila melanogaster (the fruit fly) and Mus musculus (the mouse) - produced any sort of meaningful sequence alignments. There appear to be no orthologs of IGF-1 in prokaryotes ("Insulin-like growth factor 1 (somatomedin C)").

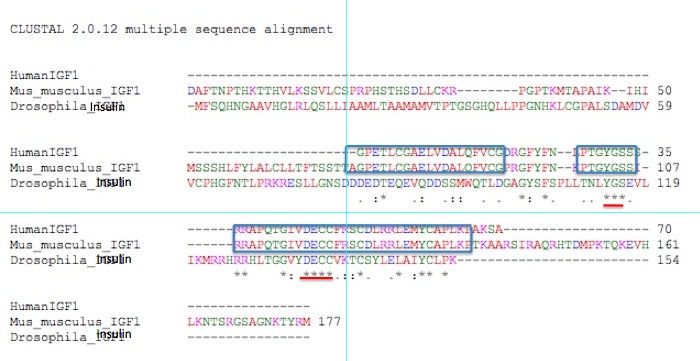
Figure 2. Ortholog of Human IGF-1 in mice (Mus musculus) and a related sequence in fruit flies (Drosophila Melanogaster).
Blue boxes indicate matching between the mouse and human version of the
protein; red underlines represent shared amino acids between all three
proteins.
Figure 2 represents the alignments of
IGF-1 ortholog protein sequences among humans, fruit flies, and mice
(the only three species out of the seven most studied in genomics). The
blue boxes indicate stretches of perfect alignment between the human
and mouse protein. These two proteins are remarkably similar in
sequence. Of the 70 amino acids comprising the human protein, residues
1-18, 20-26, 28-34, and 36-66 have perfect matches with the mouse
protein. This is a great deal of similarity.The Drosophila protein,
however, does not align so easily with either the mouse or human
version of IGF-1. The amino acid subsequences that all three organisms
share have been evolutionarily conserved, and are most likely critical
to its functioning. The portions underlined in red indicate the two
longest stretches of shared amino acids between all three proteins.
Amino acids 31-33 and 45-48 (of human IGF-1) are
the 2 longest stretches of amino acid similarity shared among these
species. It seems likely that these are necessary for the protein's
function (binding to an IGF-1 receptor protein), and that mutations in
any of these amino acids could lead to
debilitating effects. It should be noted that many biological reference sites do not include the Drosophila protein as an ortholog - the differences are simply too many ("IGF1 Orthologs"). Despite this, the Drosophila sequence can still be useful, as we have seen above.
More orthologs of Human IGF-1 need to be examined before one can truly declare that certain amino acids have been universally conserved. Zou et al. have reported the presence of an IGF-1 ortholog in zebrafish (2009). Other species with known orthologs of IGF-1 include dogs, chimpanzees, cows, rats, chickens, the African-clawed frog, and the rainbow trout ("Insulin-like growth factor 1 (somatomedin C)").
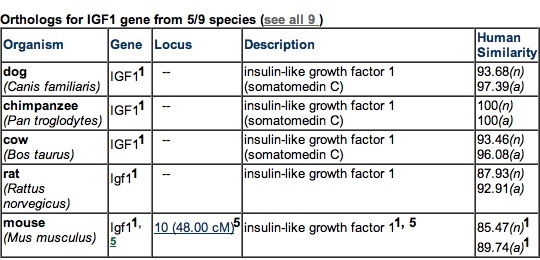
Figure 3. Five species with the most closely related IGF-1 orthologs. No prokaryotes have known orthologs of IGF-1. Permission pending from GeneCards.
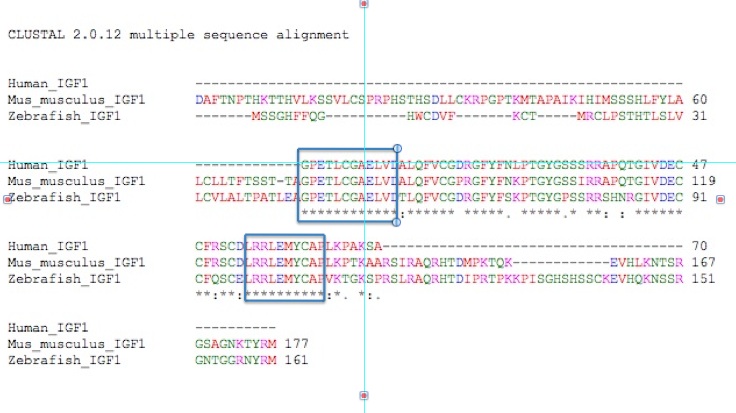
The following chart is an alignment of IGF-1 proteins from humans, mice, and zebrafish.
More orthologs of Human IGF-1 need to be examined before one can truly declare that certain amino acids have been universally conserved. Zou et al. have reported the presence of an IGF-1 ortholog in zebrafish (2009). Other species with known orthologs of IGF-1 include dogs, chimpanzees, cows, rats, chickens, the African-clawed frog, and the rainbow trout ("Insulin-like growth factor 1 (somatomedin C)").

Figure 3. Five species with the most closely related IGF-1 orthologs. No prokaryotes have known orthologs of IGF-1. Permission pending from GeneCards.
Let's take a look at some of these other
orthologs (all of which, it should be noted, occur in eukaryotes).
Analysis of more protein sequences will give us a better understanding
of what has been evolutionarily conserved. An interesting comparison
will come from analyzing a less similar protein, such as Zebrafish
IGF-1.

Figure 4. A ClustalW
alignment of IGF-1 proteins from humans, mice, and zebrafish. The blue
boxes indicate the two longest stretches of amino acid matching among
all three species.
Among the three species we are examining
here, there are some long stretches of amino acid matching. The first
12 amino acids of human IGF-1 are conserved in zebra fish and mouse
protein, indicating that these are most likely essential for IGF-1's
proper functioning. Amino acids 54-63 from the human protein are also
conserved in mice and zebrafish. These conserved amino acids most
likely play a crucial role in binding the IGF-1 receptor.
An even more expansive look at IGF-1 orthologs can be seen below:
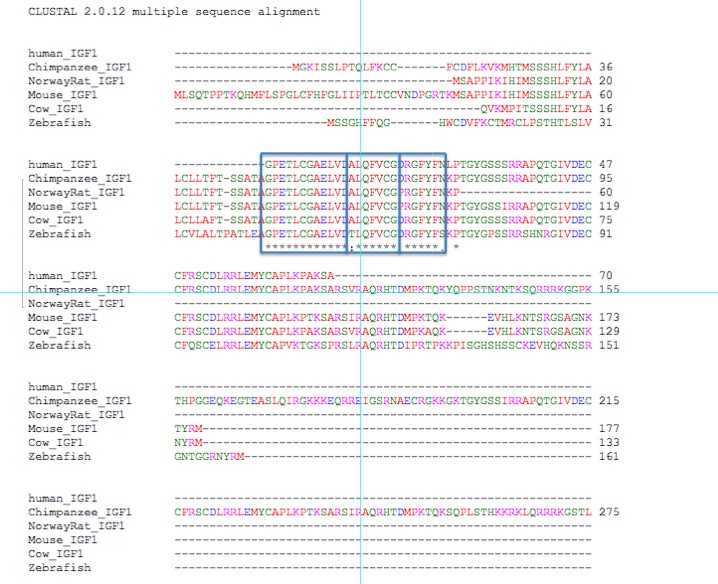
The first 27 amino acids of human IGF-1 are nearly all conserved - 25 of the 27 are conserved in each ortholog examined above. Clearly, the beginning portion of this protein is most important to its function.
An even more expansive look at IGF-1 orthologs can be seen below:

Figure 5. Alignments of 5 orthologs.
The first 27 amino acids of human IGF-1 are nearly all conserved - 25 of the 27 are conserved in each ortholog examined above. Clearly, the beginning portion of this protein is most important to its function.
Why is this important?
Examining
the protein sequences of various orthologs allows us to understand
which amino acids are most critical. With this knowledge in hand, we
can examine genetic diseases associated with mutations in the IGF-1
gene that result in differing protein sequences. IGF-1 plays a critical
role in stimulating cell growth - it is classified as a growth
stimulating hormone (a somatomedin). Mutations in IGF-1 that disrupt
binding to the IGF-1 receptor (or IGF-1R) have the potential to limit
growth. Researchers have created mutant strains of mice homozygous for
mutations in IGF-1 that either die before birth or are severely stunted
(Liu et al. 1993).
Other research with IGF-1 deficiencies/mutations has resulted in some very interesting findings. Growth hormone knockout mice (incapable of producing IGF-1) lived extremely long lives (Bartke et al. 2002). Bartke et al. propose "that the role of IGF in the regulation of growth and adult body size is important in mediating the effects of longetivity genes on aging and life span" (2002). Based on this data, mutations in IGF-1 could plausibly influence life span.
Other research with IGF-1 deficiencies/mutations has resulted in some very interesting findings. Growth hormone knockout mice (incapable of producing IGF-1) lived extremely long lives (Bartke et al. 2002). Bartke et al. propose "that the role of IGF in the regulation of growth and adult body size is important in mediating the effects of longetivity genes on aging and life span" (2002). Based on this data, mutations in IGF-1 could plausibly influence life span.
References
Bartke A., F. Dominici, D. Turyn, B.
Kinney, R. Steger, Kopchick JJ. 2003. Insulin-like growth factor
1 (IGF-1) and aging: controversies and new insights. Biogerontology 4: 1-8. You can find the abstract here.
Fulton DL., YY. Li, MR. Laird, BGS Horsman, FM. Roche, Brinkman FSL. 2006. Improving the specificity of high-throughput ortholog prediction. BMC Bioinformatics 7: 270. Find it here.
Liu JP., J. Baker, AS. Perkins, EJ. Robertson, Efstratiadis A. 1993. Mice carrying null mutations of the genes encoding insulin-like growth factor 1 (IGF-1) and type-1 IGF receptor (IGF1R). Cell 75: 59-72. Find it here.
Zou S., H. Kamei, Z. Modi, Duan C. 2009. Zebrafish IGF Genes: Gene Dupication, Conservation and Divergence, and Novel Roles in Midline and Notochord Development. PLoS ONE 4(9): e7026. Find it here.
IGF1 Orthologs. Nature: Cell Migration Consortium. 10 March 2010. Find it here.
Insulin-like growth factor 1 (somatomedin C). GeneCards. 10 March 2010. Find it here.
*Protein sequences obtained from Blast. Sequences were found after searching with human IGF-1*
*All alignments were done using ClustalW*
Fulton DL., YY. Li, MR. Laird, BGS Horsman, FM. Roche, Brinkman FSL. 2006. Improving the specificity of high-throughput ortholog prediction. BMC Bioinformatics 7: 270. Find it here.
Liu JP., J. Baker, AS. Perkins, EJ. Robertson, Efstratiadis A. 1993. Mice carrying null mutations of the genes encoding insulin-like growth factor 1 (IGF-1) and type-1 IGF receptor (IGF1R). Cell 75: 59-72. Find it here.
Zou S., H. Kamei, Z. Modi, Duan C. 2009. Zebrafish IGF Genes: Gene Dupication, Conservation and Divergence, and Novel Roles in Midline and Notochord Development. PLoS ONE 4(9): e7026. Find it here.
IGF1 Orthologs. Nature: Cell Migration Consortium. 10 March 2010. Find it here.
Insulin-like growth factor 1 (somatomedin C). GeneCards. 10 March 2010. Find it here.
*Protein sequences obtained from Blast. Sequences were found after searching with human IGF-1*
*All alignments were done using ClustalW*
Davidson College
Davidson Molecular Biology 2010 Home
Michael Rydberg's Molecular Home
Please contact me with any comments or questions at mirydberg@davidson.edu