This web page was produced as an assignment for an undergraduate course at Davidson College
Red Fluorescent Protein
Introduction
Red fluorescent protein (RFP) is a protein commonly used to examine intercellular processes such as tracking gene expression and tagging other proteins. The original protein, isolated from Discosoma coral, is a tetramer commonly referred to as dsRed. DsRed is not very useful in lab applications because it has a very slow maturation rate (~12 h) and its tetramer structure could result in unwanted associations of proteins (Strongin et al. 2007). Therefore, several synthetic variants of the protein have been created, including monomer variants that are much smaller and mature faster. However, for the purposes of this website we will examine the wild type protein (dsRed) and refer to it as RFP.
Function
RFP adds to the natural coloration of cells, typically creating a red hue. The fact that it fluoresces allows for many applications in the scientific community as discussed above. Yet despite its usefulness, the natural function of dsRed is not clearly understood. A few possible theories have been put forward claiming it may have some function in protection against UV light, protection against predators, or that it may be linked to successfully competing against other coral (Dove et al. 1995). Regardless, RFP’s basic function is to emit light (maximum of 583nm) in response to a specific light wavelength (maximized at 558 nm) (Gross et al. 2000).
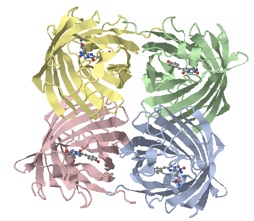
Structure
RFP consists of two eleven-stranded beta barrels of identical 234 amino acid sequences pairing into a dimer that associates into a tetramer (Yarbrough et al. 2001). Under certain circumstances, two of these tetramers may also weakly associate to form an octamer (Baird et al. 2000). In the tetramers, the subunits are held together by amino acid interactions, by hydrophobic interactions, and by hydrogen bonds (Bergmann 2004).
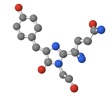
Click here for RFP’s amino acid sequence (NCBI)
How Structure Relates to Function
As with most proteins, the exact positioning of certain amino acids within RFP is extremely important to how the protein functions. For example, the methylation of Lys83 causes the emission maximum of RFP to shift to 602 nm; also, the mutation of Lys83 to Arg causes to the protein to halt at the green stage of its maturation (Baird et al. 2000). It is also interesting to note that RFP folds in a similar manner to green fluorescent protein (GFP) despite the fact that both their amino acid sequences share roughly 23% homology (Yarbrough et al. 2001). This could potentially demonstrate evolution conserving a structure from a common ancestor as organisms differentiated.
Additionally, the location of the chromophore contributes strongly to RFP’s function. The chromophore’s location allows for specific interactions with surrounding amino acids that create and maintain an electrical charge within the chromophore (Bergmann 2004). This is significant because chromophores work by light hitting the chromophore and exciting the chromophore’s electrons. Then, these electrons emit light of a particular wavelength as they settle back to their normal state (PHYSorg.com c2006). It is essential that the chromophore’s charge is maintained if it is to be responsive to a particular wavelength of light, because the light energy is a precise amount of energy in a narrow range of energy levels that can excite the electrons of the specific chromophore. If the chromophore’s charge were altered, it would take a completely different amount of light energy (different wavelength) to excite the electrons and completely change the specificity of the protein’s interactions with the outside world.
References
Baird GS, Zacharias DA, Tsien RY. 2000. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA [Internet]. [cited 2010 Feb 15];97(22):11984-11989. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC17281/?tool=pubmed
Bergmann R. 2004. Red Fluorescent Protein from a Discosoma coral. University of Hamburg [Internet]. [cited 2010 Feb 15]. Available from: http://www.biologie.uni-hamburg.de/lehre/bza/light/1g7k/1g7ktext.htm
Dove SG, Takabayashi M, Hoegh-Guldberg O. 1995. Isolation and Partial Characterization of the Pink and Blue Pigments of Pocilloporid and Acroporid Corals. Biol Bull [Internet]. [cited 2010 Feb 15];189:288-297. Available from: http://www.biolbull.org/cgi/content/abstract/189/3/288
Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. 2000. The Structure of the Chromophore within DsRed, a Red Fluorescent Protein from Coral. Proc Natl Acad Sci USA [Internet]. [cited 2010 Feb 15];97(22):11990-11995. Available from: http://www.jstor.org/stable/123778
Strongin DE, Bevis B, Khuong N, Downing ME, Strack RL, Sundaram K, Glick BS, Keenan RJ. 2007. Structural rearrangements near chromophore influence the maturation speed and brightness of DsRed variants. Protein Engineering, Design & Selection [Internet]. [cited 2010 Feb 15];20(11):525-534. Available from: doi:10.1093/protein/gzm046
Jumpy electrons make chromophores semiconductors suitable for nanoscale electronics. [Internet]. PHYSorg.com; c2006 [cited 2010 Feb 15]. Available from: http://www.physorg.com/news70896830.html
Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. 2001. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-Å resolution. Proc Natl Acad Sci USA [Internet]. [cited 2010 Feb 15];98(2):462-467. Available from: http://www.jstor.org/stable/3054705
Image Sources
-In order of appearance-
Image 1 - FPbeachTsien.jpg from Wikipedia. Distributed under GNU Free Documentation License,
Image 2 - RFP, DsRed Molecule. Generated via pdb.org and Jmol.
Image 3 - Chromophore molecule. Generated via pdb.org and Jmol.
Image 2: RFP
Image 3:
Chromophore
Image 1: Fluorescent Bacterial Colonies