This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Proteins!
Annotated Gene: CCL1/YPR025C (YPD Protein Report) Brief report of YPR025C Microarray data for YPR025C
Annotated Gene: CCL1/YPR025C (YPD Protein Report)
Saccharomyces cerevisiae Chromosome XVI
SGD: 50006229 Genbank: CAA89279.1 MIPS (PIR): 539383 SWISS-PROT: P37366
While the microarray data for CCL1 shows this gene clustering with many varied annotated and non-annotated genes under several different experimental conditions, the field of proteomics has also been making its contribution to the discovery and annotation of the way cells work. Although disjointed, the various protein databases have expanded upon the information gathered by microarrays. For example, the Database of Interacting Proteins shows six other proteins that interact with the protein cyclin CCL1, most of which did not appear linked in previously data:
![]() Retrieves
information about a particular interaction
Retrieves
information about a particular interaction
![]() Retrieves
1 level of interaction links.
Retrieves
1 level of interaction links.
(table courtesy of DIP, permission requested)
The Yeast Protein Database agrees with the table above that KIN28 and TFB3 are both linked to CCL1, but also relates VID22. A yeast integration circuit showing protein interactions published by Schwikowski, Fields, and Uetz (shown below) also connects CCL1 with KIN28, but also shows lines to RAD59 and SNZ3, which are linked by YPD to each other but not to CCL1 or KIN28.
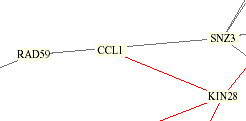
KIN28, as a subcomponent of the kinase sub complex TFIIK of TFIIH along with CCL1, has been a known interaction. The red line above indicates that their cellular and subcellular localizations are identical. Most of the other proteins are involved with DNA repair with the exception of VID22, which is only known to be in the plasma membrane, and SNZ3, which is connected to metabolism and cell stress, as well as involved with the biosynthetic enzyme vitamin B6. Ito et al. (1), who are cited by the YPD, used the two-hybrid method and compared their results with Uetz et al. (2) and found that not much overlap occurred, resulting in greater knowledge of the yeast proteome interactions.
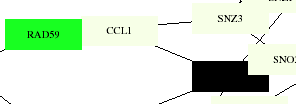
This membrane protein interaction map shows CCL1 connected with the same proteins as the larger scale one, with RAD59 color-coded as a cell cycle control protein, which makes sense as one of its roles is in meiosis. The black box represents KIN28, which is coded for RNA turnover. The aging map codes both CCL1 and KIN28 as pol-II transcription factors, which they have been identified as by the YPD as well. The degredation map shows no particular color distinction for any of these proteins.
The Yeast Resource Center links CCL1 with PRP16, as the table above does; PRP16 is an RNA binding protein that also plays a role in RNA splicing. The PathCalling Yeast Interaction Database claims no interactions for CCL1. The Triples database (3, 4) disrupted a CCL1 clone with the mTn transposon and the protein was still expressed, which means that the insertion randomly landed downstream of the promotor. The subcellular location was unable to be determined by this database because it failed to stain above a background level.
Databases consulted with no significant results for CCL1:
Enzymes and Metabolic Pathways Database
Non-Annotated Gene: YHR078W (YPD Protein Report) Brief report of YHR078W
S. cerevisiae Chromosome VIII
SGD: S0001120 Genbank: AAB68887.1 MIPS (PIR): S46809 SWISS-PROT: P38799
Besides the microarray data for this gene, not much information exists even in protein databases. WIT links YHR078W with integral nuclear envelope protein NDC1, which is an annotated gene. Badcock and Churcher say here that this protein (assumably NDC1) is essential for mitotic viability as it is required for spindle pole body duplication in G1 and may insert the spindle pole body into the nuclear envelope. They also posit the ER as a possible localization (5). NDC1 functions as a nuclear export/import protein, which correlates with the previous microarray data found.
In light of this data, a possible experiment to find out more information and see if YHR078W is actually involved in transport as predicted would be the yeast-two hybrid method. Since no other genes have been linked to this one besides the one that codes for NDC1, it would be a logical choice as prey. Other possibilities include the products of the genes from the microarray data that clustered with YHR078W and were involved in transport, such as BOS1, GEA1, FTR1, MSP1, and AGP3. Depending on what it linked with, it would be possible both to verify YHR078W function and to solidify its subcellular localization. Since so little information about this particular gene and resulting protein actually exists, any positive results with this experiment could possibly result in a breakthrough for YHR078W.
Databases consulted with no significant results for YHR078W:
Database of Interacting Proteins
Schwikowski, Fields, and Uetz PDF Files:
NB_Figure1 Membrane Aging Degredation
PathCalling Yeast Interaction Database
References
1. Ito, Takashi, Tomoko Chiba, et al. 10 April, 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA, Vol. 98(8):4569-4574. 8 November, 2001. http://www.pnas.org/cgi/content/full/98/8/4569
2. Uetz, P., L. Giot, et al. 10 February, 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature, Vol.403(6770): 623-7. 8 November, 2001. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10688190&dopt=Abstract
3. Kumar, A., Cheung, K.-H., Ross-Macdonald, P., Coelho, P.S.R., Miller, P., and Snyder, M. (2000). TRIPLES: a Database of Gene Function in S. cerevisiae. Nucleic Acids Res. 28, 81-84. (Full-text in PDF reproduced with permission from NAR Online http://www.oup.co.uk/nar )4. Ross-Macdonald, P., Coelho, P.S.R., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K.-H., Sheehan, A., Symoniatis, D., Umansky, L., Heidtman, M., Nelson, K., Iwasaki, H., Hager, K., Gerstein, M., Miller, P., Roeder, G.S., and Snyder, M. (1999). Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402, 413-418. Nature 402, 413-418.
5. Badcock, K., C. Churcher, et al. Submitted (Nov 1994) to EMBL/Genbank/DDBJ Data Banks. http://wit.mcs.anl.gov/WIT2/CGI/prot.cgi?user=&org=SC&prot=sp|P32500