
"This web page was produced as an assignment for an undergraduate course at Davidson College."
Expression of YPR098C
The Expression Connection database is part of the Saccharomyces cerevisiae Genome Database (SGD) webservice and it provides access to eleven different DNA microarray experiments. When I typed in the ORF name, YPR098C, I obtained the following results.
This is the scale used in all of the experiments to show gene expression and repression:

Figure 1. Scale depicting color scheme for gene expression analysis of DNA microarray experiments. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
The first experiment I studied was the response of the yeast genome to different alpha-factor concentrations. Alpha-factor is a mating pheromone that arrests the cell cycle of yeast at the G1 phase and induces genes involved in mating (Roberts et. al.). Therefore, genes involved in any way in the mating response, such as shape change, movement, etc. should be induced in this DNA microarray experiment. The expression pattern of YPR098C is shown below:
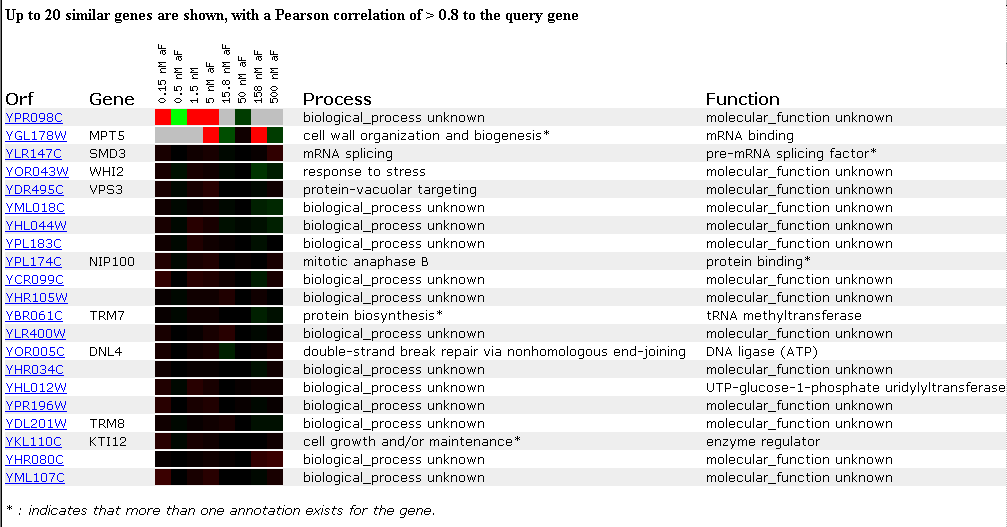
Figure 2. Expression at different alpha-factor concentrations. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
YPR098C has an interesting expression pattern in this experiment. It is induced at the lowest concentration of alpha-factor (.15nM), repressed at the next concentration (.5nM), induced again at 1.5nM and 5nM, and is finally repressed at a high concentration of alpha-factor (50nM) (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl). Unfortunately no data was obtained for the higher concentrations, 158nM and 500nM, so it is difficult to interpret the pattern of gene expression. Most of the genes that clustered with the YPR098C Open Reading Frame (ORF) show no change in expression throughout the duration of the experiment. The YGL178W ORF shows the most similar expression pattern, although the pattern isn't identical, because it is similarly turned on and off throughout the experiment. The gene associated with this ORF, MPT5 is known to function as an mRNA binding protein involved in cell wall organization, biogenesis, and re-entry of cells into the mitotic cell cycle after pheromone arrest. Therefore, although this gene is located on a different chromosome, chromosome VII, using "guilt by association," this experiment suggests that the YPR098C ORF might code for a protein involved in preparing the cell for mating in response to the alpha-factor pheromone, since transcription of this ORF is induced at low and medium levels of alpha-factor.
Interestingly, another experiment in this database also used exposure to alpha-factor as the experimental condition, but this DNA microarray was looking at the effect of alpha-factor exposure over time rather than at different concentrations. The DNA microarray results are below:
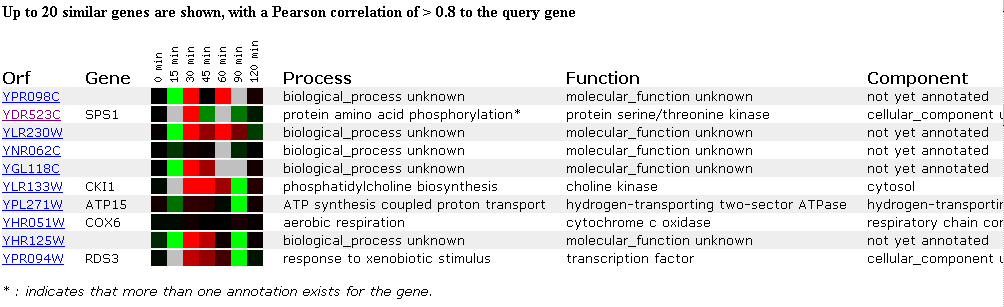 Figure 3. Expression
in response to alpha-factor. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Figure 3. Expression
in response to alpha-factor. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
As seen in the previous experiment, when the Saccharomyces cerevisiae cells were exposed to different alpha-factor conditions, the expression of the YPR098C ORF was affected. Small amounts of alpha-factor caused this ORF to be induced in the previous experiment, while in this experiment, the expression of YPR098C is repressed when exposed to alpha-factor for a short time (15 minutes). After 30 minutes of exposure to alpha-factor, the YPR098C ORF becomes induced, and the expression level returns to a normal state after 45 minutes. After 60 minutes of exposure to alpha-factor, the gene is again induced and returns to normal expression levels after 120 minutes. This expression pattern can also be depicted in the form of a graph:
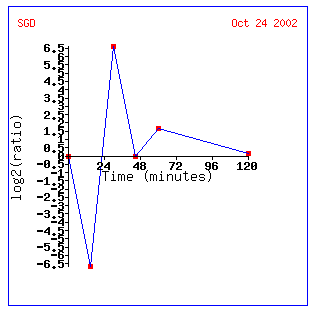
Figure 4. Logarithmic graph depicting the results from the DNA microarray experiment testing the expression of the YPR098C ORF in response to alpha-factor. The exposure time to alpha-factor is on the x-axis and the log2(ratio) of expression is on the y-axis. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Unfortunately, the ORF's that the YPR098C ORF most closely clustered with also have unknown functions and so not much information is gained about the role of YPR098C in the yeast cell. The gene SPS1 did have a similar expression pattern and is a kinase involved in protein phosphorylation. It is logical that this gene is induced and then repressed as the genome is exposed to alpha-factor for increasing amounts of time, because it is involved in regulating mitosis and meiosis, cell cycles that will need to be activated as the cell undergoes the mating process. The results of this experiment seem to suggest that the YPR098C ORF encodes a protein that is a kinase that functions to regulate some aspect of the mating process by phosphorylation, but since I previously concluded that this ORF would encode a membrane protein, further evidence is needed before a more definate conclusion can be made regarding the functional role of the YPR098C ORF in the cell. Another clustered gene with a known function is ATP15, which is involved in ATP synthesis coupled proton transport, but since the 90 minute time point data is not available for the YPR098C ORF, it is difficult to compare the expression of these two DNA sequences.
Click here to read the scientific paper that produced the above data sets on the effect of alpha-factor on yeast genes.
In this next experiment, gene expression was monitored as cells switched from anaerobic to aerobic functions upon depletion of their glucose sources and thus, the cells underwent a metabolic switch from fermentation to respiration. As seen below, the YPR098C ORF is induced after 18.5 hours under aerobic conditions. Most genes that show this pattern of induction when placed in an aerobic environment encode various enzymes or proteins involved in respiration. The other genes in this cluster have different roles such as YGP1, which is located in the cell wall and responds to stress and HAP4 which is a transcriptional activator. Due to the diverse nature of genes displaying similar expression patterns, it is difficult to conclude much from this experiment, but obviously, the protein that the hypothetical ORF YPR098C encodes is somehow involved in fundamental cellular processess that occur during aerobic respiration since it is induced after a long period of aerobic respiration.
Click here to read the full paper on this experiment.

Figure 5. Expression during the diauxic shift. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)

Figure 6. Graph depicting the DNA microarray data of YPR098C ORF expression during the diauxic shift. The aerobic respiration time is plotted on the x-axis, while the log2 (ratio) of expression is on the y-axis. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
In another experiment, gene expression was observed in response to varying zinc levels. Zinc is an essential nutriet for cells and has many functions, but the method of its uptake and the genes affected by its concentration remain unknown. In this experiment, gene expression in wildtype cells, both replete and deficient of zinc, were compared to gene expression in zap1 mutants. Zap1 is a transcription factor which senses zinc dificiency and subsequently increases the expression of certain target genes.
The expression pattern of the YPR098C ORF was most similar to the known genes ATP17 and HMRA2. ATP17 encodes a protein involved in ATP synthesis coupled proton transport and HMRA2 encodes a protein involved in mating-type specific transcriptional regulation. Expression of the YRP098C ORF was repressed in the wildtype cells deficient of zinc and decreased as the zinc concentration increased in the zap1 mutants. This suggests that the YPR098C ORF is not a target of zap1p. It is also interesting to note that many genes involved in aerobic respiration are clustered with the YPR098C ORF in this experiment since this ORF had induced expression in the previous experiment where the conditions were switched from anaerobic to aerobic conditions. Maybe this ORF encodes a protein that is involved in aerobic respiration. Further experiments must be analyzed and performed before this conclusion can be made.
Click here to read the paper published on this experiment.
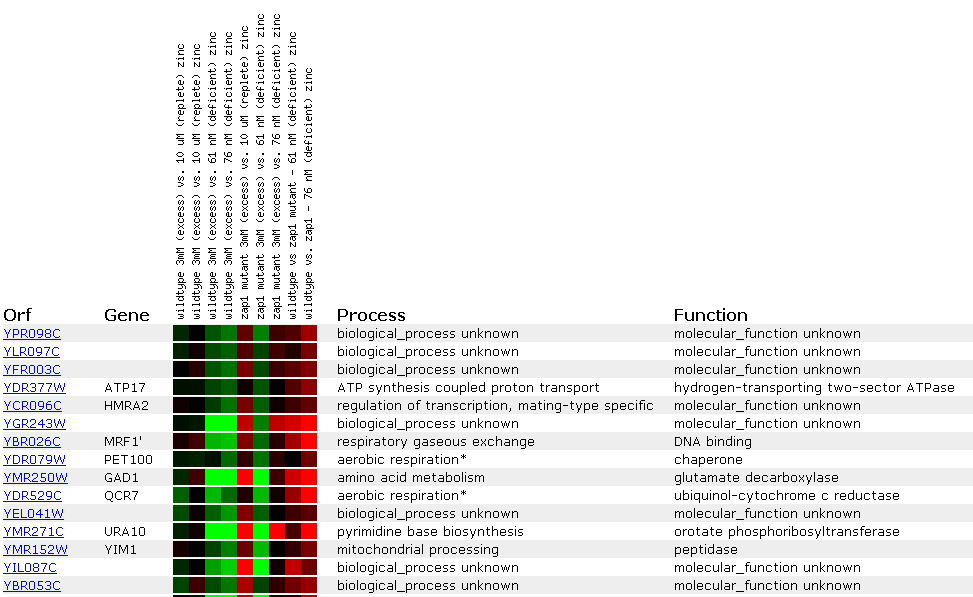 Figure 7. Expression in response
to varying zinc levels. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Figure 7. Expression in response
to varying zinc levels. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Click here to see the rest of this DNA microarray data. The data was too long to fit onto one screen.
Another experiment was performed to examine the yeast genome response to DNA-damaging agents. Many proteins in the cell monitor genomic integrity and are activated when DNA damage is detected. The response activated includes cell cycle arrest, altered gene expression, DNA damage repair, and cell death. The cell doesn't want damaged DNA to be replicated and passed on. Many kinases help regulate the DNA detection and damage response. Mec1, dun1, and crt1 are genes that encode such kinases and therefore, mutants lacking these genes are hypersensitive to DNA damage since part of their detection and repair pathway is mutated.
Some interesting data was obtained for the YPR098C ORF from this experiment. Although the ORF didn't cluster with any other ORF's, gene expression differences were observed between experimental conditions and normal conditions. Slight induction of the YPR098C ORF was observed when methylating agents were exposed to the wildtype cells for a lengthy time period, but no other expression changes were observed in response to methylation. Slight induction of this ORF was also observed when the wildtype cells were exposed to irradiation, but no expression changes were observed when mec1 mutants were exposed to irradiation. Repression of this ORF was observed when mec1 mutants were treated with the mock irradiation conditions although no expression changes were observed when wildtype cells underwent this treatment. I thought the most interesting data from this experiment resulted from the exposure to heat experiments. When wildtype cells were exposed to heat (37 degrees Centigrade), the YPR098C ORF was strongly induced. This ORF was also induced when mec1 and dun1 mutants were stressed with heat. This data suggests a possible involvement of the YPR098C hypothetical protein in responding to cellular stresses such as irradiation and heat. Further analysis is needed to make more substantial claims.
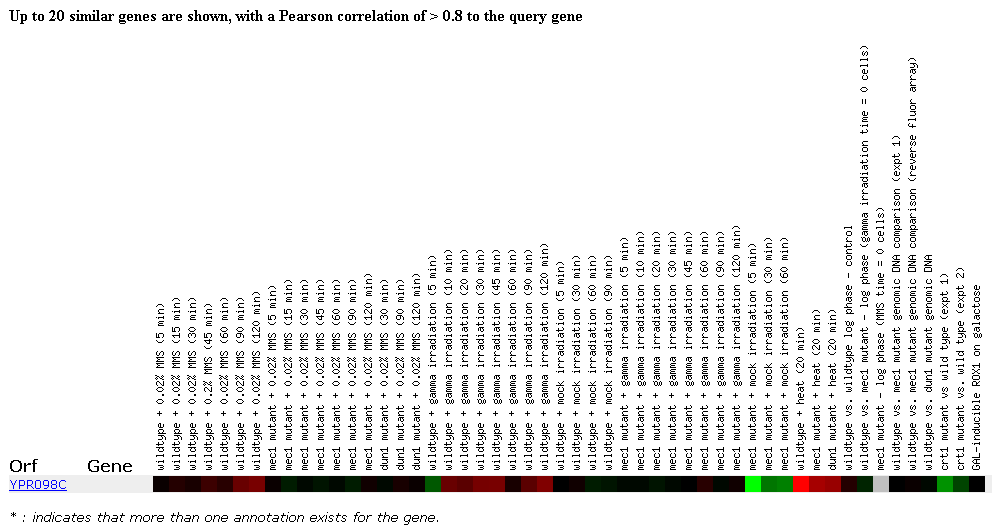
Figure 8.Expression in response to DNA-damaging agents. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Click here to read the paper detailing this DNA-damage experiment.
The final experiment that I'm going to examine involves observing the effects of MANY environmental changes on the yeast genome. These environmental changes include various methods of heat shock, the addition of sorbital, hydrogen peroxide, DTT, diamide, or menadine, hypo-osmotic shock, amino acid starvation, nitrogen depletion, a diauxic shift, stationary phase, gene overexpression, exposure to sugar, and steady state experiments. This is an impressive number of experiments for one group to perform and although it provides a large amount of data, I didn't learn that much from the other ORF's that YPR098C was clustered with. Expression of the YPR098C ORF was induced in the heat shock, hydrogen peroxide, diamide, nitrogen depletion, and stationary phase experiments, and was repressed in the hypo-osmotic shock and glucose experiments so this is where I will focus my attention.
The greatest amount of induction of the YPR098C ORF was observed when the cells were heat shocked for 20 minutes beginning at different temperatures, but all reaching the final temperature of 37 degrees celsius. Induction was the greatest when the cell underwent the greatest temperature changes as seen below. Induction of this ORF was also high after a 40 minute exposure to hydrogen peroxide, but the induction did not increase linearly as the exposure time was increased. When the cells were exposed to DTT, induction increased linearly with time of exposure, but induction peaked at 50 minutes out of 90 minutes during exposure to diamide. Induction also increased respectively as nitrogen was depleted and metabolism was shifted from anaerobic to aerobic conditions. Induction was also strong in the stationary phase experiments.
Unfortunately, the YPR098C ORF clustered again with many genes with unknown functions. It did cluster with a few genes involved in protein synthesis and response to oxidative stress and after observing the DNA microarray data, it seems that the YPR098C ORF might play a role in responding to oxidative stress.
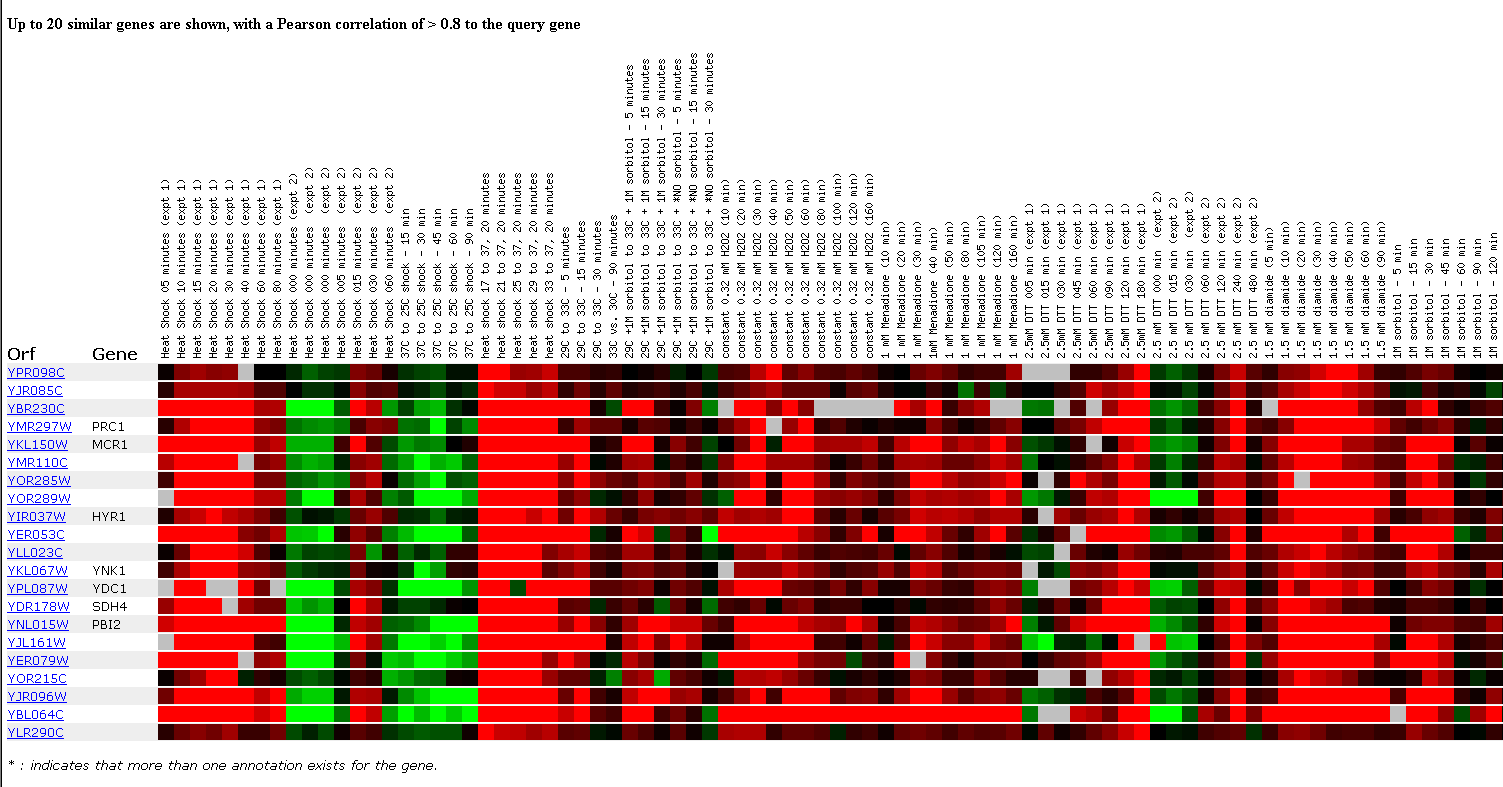
Figure 9. Expression in response to environmental changes. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
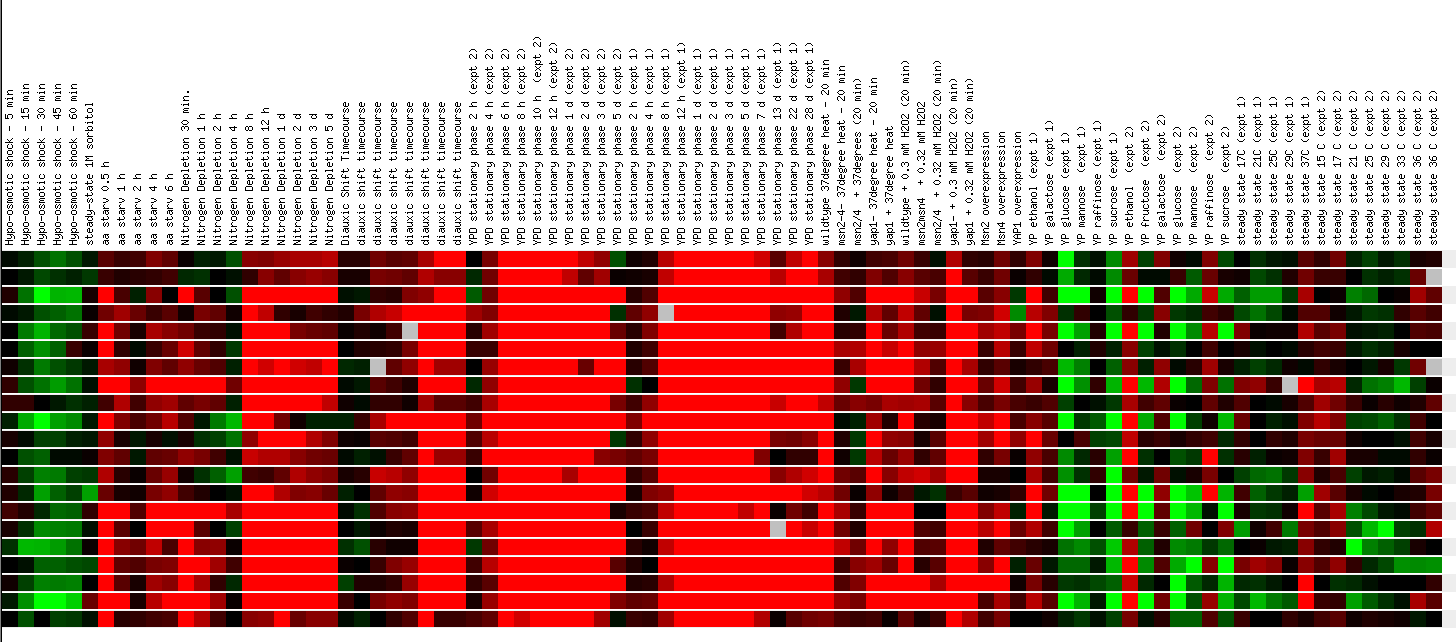
Figure10. Continued Expression in response to Environmental changes. The second half of the experiments performed with the genes in the same order as in figure 9. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
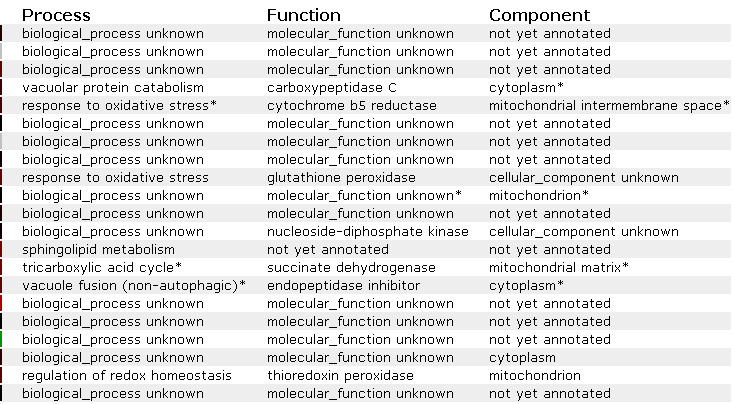
Figure 11. Process, function, and component information for the genes in the previous experiment that are depicted in figures 9 and 10. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Click here to read the paper explaining this environmental change experiment.
Additional information can also be obtained at another SGD website named Function Junction. The yeast microarray global viewer section of this webpage provides data from a large number of experiments involving the specific gene of interest entered by the user. This information can be very helpful, but not much information is included about the experimental conditions, so sometimes the results are difficult to interpret. Therefore, I only obtained general information from this site. Much more specific conclusions and predictions would be able to be made after investing this site further.
Interestingly, in the first experiment (Angus-hill--Rsc3-Rsc30), YPR098C is initially induced, then repressed, then induced again. This experiment involves mutant chromatin remodeling complexes and I would be interested to learn more about the experimental conditions that produced the results shown in figure 12. Click here to read the abstract for this paper. Compared to my known gene, FHL1, I was surprised to find that the hypothetical ORF YPR098C exhibited greater changes in expression in the majority of the DNA microarray experiments. Therefore, although YPR098C is a small ORF, it appears to encode a functional protein. In glancing at the experiments on the Function Junction webpage, I noticed that the YPR098C ORF was induced in experiments involving: MAPK, metabolism, mitochondria, rapamycin, stress, MMS, and Rpn4. These results agree with the results obtained from Expression Connection. It seems that YPR098C plays some role in stress response connected to possibly the mating response or metabolism. Expression of the YPR098C ORF remained normal in the Mec1 mutant, mitotic, sporulation, aneuploidy, anaerobic respiration, cell cycle, and fork head experiments. Expression of the YPR098C ORF was repressed in the salt and UPR experiments.
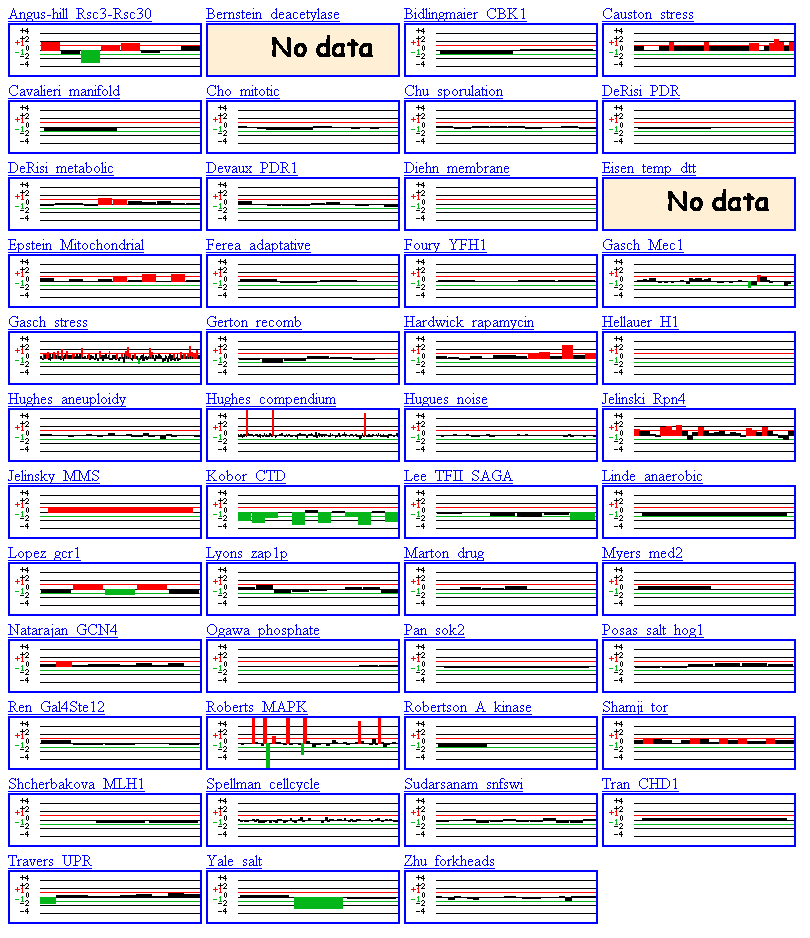
Figure 12. DNA Microarray experiments performed involving the YPR098C ORF. Summaries of the results are observed in the small rectangles under the name of the experiment or researcher. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/functionJunction)
Click here to access the Function Junction Webpage
Conclusions: In preparing this webpage, I looked through a large number of DNA microarray data sets. Although many unknowns still remain in terms of unknown gene functions, the availability of public databases such as these will help uncover new information about such genes with unknown functions. In assignment 2 for this genomics class I used sequence analysis and predictive tools in an attempt to gain information about the YPR098C ORF located on Saccharomyces cerevisiae chromosome XVI. The only information that I gained was that this ORF probably encodes a membrane protein. The DNA microarray data information that I searched allowed me to observe how and under what conditions the YPR098C ORF is expressed and provided some clues to its biological role. My conclusions from this data are only hypotheses about the function of the YPR098C ORF, but it appears to me that this ORF encodes a functional membrane protein involved in cell wall organization that is involved in helping prepare the cell for mating and for dealing with stresses such as heat shock and DNA-damage. Since I'm not an expert on yeast, I don't know if this combination of roles is possible for one small ORF, but the microarray data suggests that this ORF at least has some of these functions. I will be interested to investigate further experiments performed on this DNA sequence, which is surprisingly very different in gene expression from one of its neighboring genes, FHL1. I would be especially interested in observing gene expression in various YPR098C mutants under the same experimental conditions.
References:
DeRisi JL, et al. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278(5338):680-6.
Lyons TJ, et al. (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. PNAS 97(14):7957-62.
Gasch AP, et al. (2001) Genomic expression responses to DNA-damaging
agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell
12(10):2987-3003.
Gasch AP, et al. (2000) Genomic expression programs in the response of
yeast cells to environmental changes. Mol Biol Cell 11(12):4241-57.
Send comments, questions, and suggestions to: sahenry@davidson.edu