This web page was produced as an assignment for an undergraduate course at Davidson College.
Gene Expression of Gln3 and YEN1
I will describe the gene expression in the annotated gene, Gln3, and the non-annotated gene YEN1. Using DNA microaaray data, gene expression and analysis is thorough and extremely informative.
Gln3 Expression
As mentioned in the second web assignment, Gln3 gene encodes a transcription factor (Gln3p) found in the nucleus and cytoplasm which plays a role in nitrogen catabolite repression (NCR). S. cerevisiae preferentially uses good nitrogen sources instead of bad ones "by repressing GATA-factor dependent transcription of the genes needed to transport and catabolize poor nitrogen sources"(Cox, et al., 2000, http://genome-www.stanford.edu/Saccharomyces/yeast00/abshtml/091.html).
Using Expression Connection, eleven databases were searched which gave me results of DNA microarrays under different experimental conditions. Gln3 expression for some microarrays produced little to no change in induction or repression, therefore, I will focus my discussion on Gln3 expression which pertains most to its biological process and molecular function in the cell. Below is a list of the different experimental conditions used to create DNA microarray data for Gln3.
Expression at different alpha-factor concentrations (Rosetta
Inpharmatics)
Expression in response to alpha-factor (Rosetta Inpharmatics)
Expression in response to DNA-damaging agents (Stanford University)
Expression during the diauxic shift (Stanford University)
Evolution of expression during glucose limitation (Stanford University)
Expression Regulated by the PHO pathway (Stanford University)
Expression in response to environmental changes (Stanford University)
Expression during the cell cycle (Stanford University, Cold Spring Harbor)
Expression in response to histone depletion (The Whitehead Institute)
Expression during sporulation (USF, Stanford University)
Expression in response to varying zinc levels (University of Missouri, Stanford
University)
Expression in response to environmental changes for GLN3
One of the most interesting microarrays concerned the expression in response to environmental changes. Since Gln3 regulates nitrogen metabolism, we would expect to see changes in expression as the environmental nitrogen and other nutrients change.
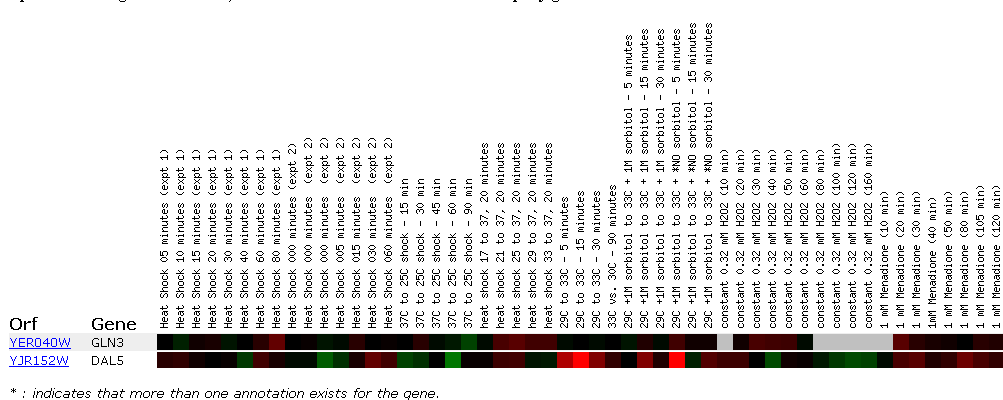

This data reinforces the molecular function of GLN3. As nitrogen is depleted, Gln3 is inducted almost 3 fold. This causes the shorter mRNA transcript to be produced from NCR sensitive genes (genes sensitive to quality and quantity of nitrogen in the environment). The short mRNA's production utilizes "bad" or low nitrogen sources, which in essence is what is happening when nitrogen is depleted in the microarray. Repression (green) or no detectable signal (black) means that nitrogen is not a limiting factor in the other environmental stresses shown in the array. Dal5 is an allantoate transporter in the plasma membrane and facilitates the movement of this molecule in and out of the cell. Below is a picture of allantoate. Inferring from the microarray when nitrogen sources are depleted this transporter is induced to facilitate the movement of allantoate into the cell.
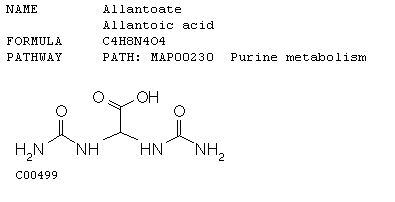
This image was obtained from http://www.biologie.uni-hamburg.de/b-online/kegg/kegg/db/ligand/cpdhtm/C00499.html
Expression in response to histone depletion
Histones are a group of proteins found in chromosomes with the DNA that are thought to play a role in cellular metabolism and gene control.They have numerous and distinct types due to varying methylation, acetylation, and phosphorylation, but can be generalized by having high levels of lysine and arginine.( http://www.worthington-biochem.com/manual/H/H.html). In keeping with Gln3, the depletion of histones seems to have an effect with regard to the ability for a yeast cell to metabolize nitrogen successfully.
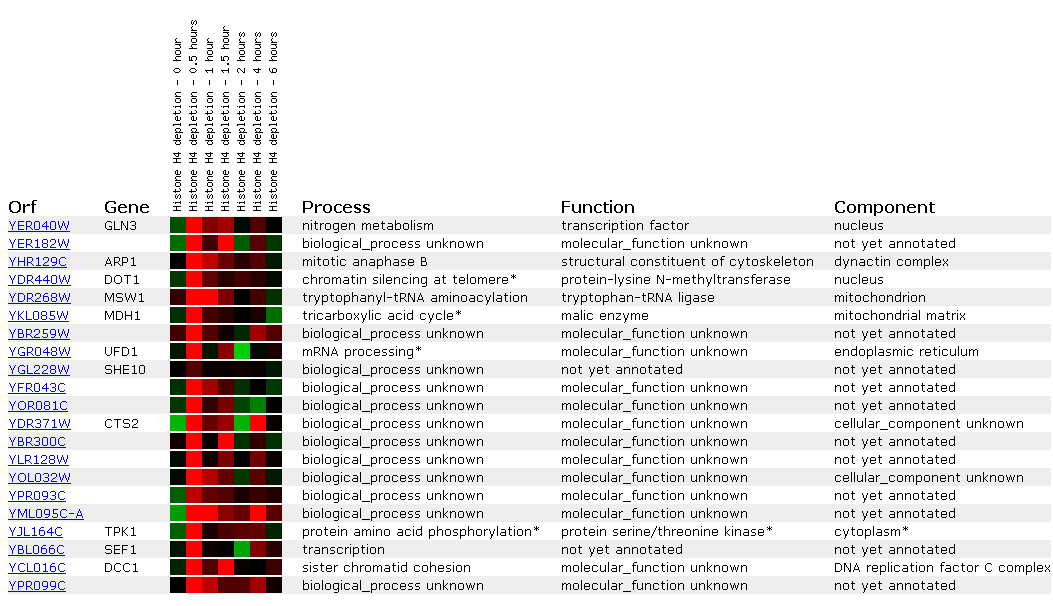
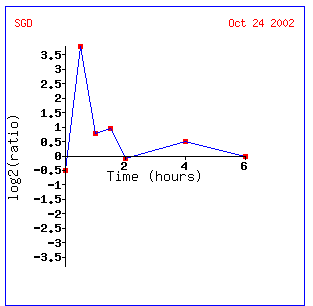
Within one half hour of depletion, Gln3 is induced. However, induction is quitted after an hour. This may occur because the levels of small mRNA's needed to regulate the metabolism of nitrogen is sufficient, or Gln3 may be a transcription factor for some other gene. The other genes most like expression seen in Gln3 have unknown biological processes. TPK1 and DOT1 appear similar. DOT1 is involved in histone methylation and silencing, while TPK1 is a phosphorylator. Both are regulatory genes.
Expression in response to alpha-factor for GLN3
Alpha-factor is a pheromone which induces genes necessary for mating. It impedes yeast in G1 of the cell cycle. It can also cause morphological changes (http://www.zymor.com/y1001.html). Thus it is not out of the question that many biological processes stop or slow down in order for mating to occur, in this case, nitrogen catabolism.
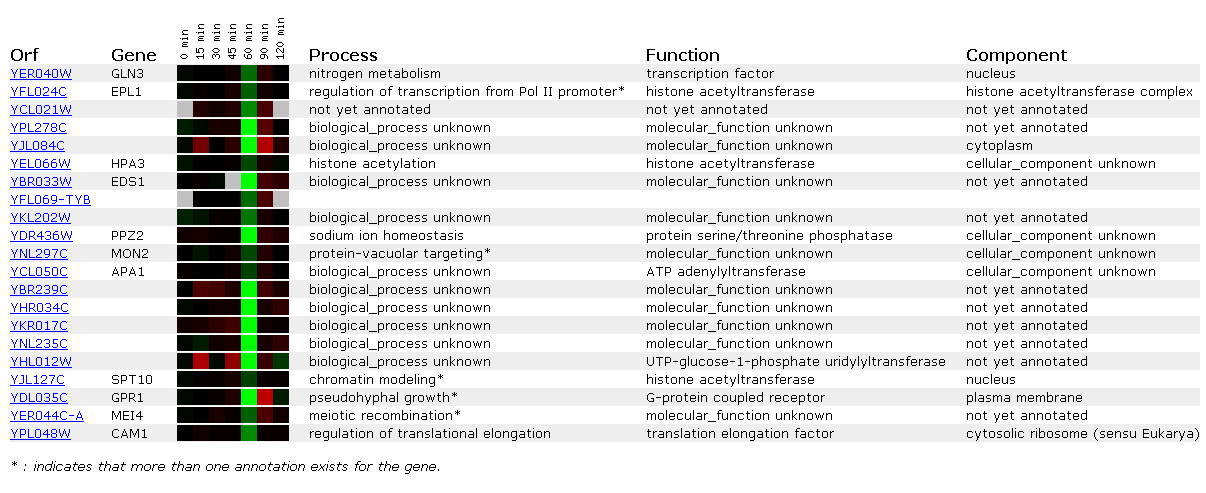
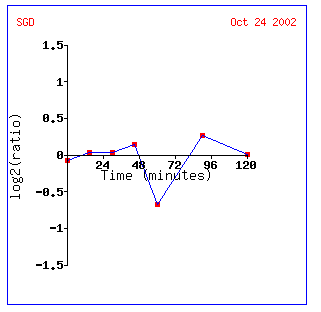
Gln3 is repressed about .85-fold at the one hour mark. Interestingly all the other genes repressed in the figure occur at the one hour mark. The delay might be cause by a need for a certain concentration of alpha-factor in order to initiate mating protocols. EPL1 also regulates transcription and the other genes are regulators of cellular growth and diversity.
Expression during the diauxic shift for GLN3
Diauxic shift is the transition from anaerobic to aerobic respiration. Gln3 was not impacted very much by the switch. It was slightly repressed then slightly induced but overall, not affected very much. Nitrogen metabolism increased with the addition of oxygen. However, oxygen does not have much bearing on the concentration of nitrogen in the system, thus will not affect the induction/repression of Gln3.
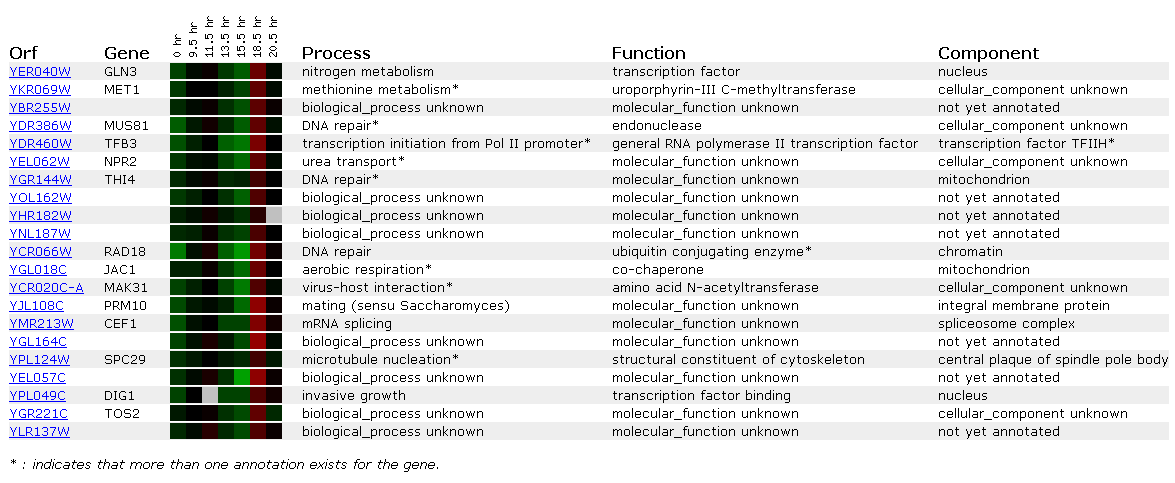
However, if you look at genes with similar expression patterns, they appear important in the shift from anaerobic to aerobic respiration. NPR2 is involved in urea transport, while JAC1 is involved in aerobic respiration itself! Why not a huge induction? JAC1 actually consumes oxygen so that levels don't become toxic to the cell; it actually is inviable without the protein(SGD, http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=YGL018C) . We most likely don't see a huge induction for this reason. A yeast cell would not deplete itself of a vital protein even under anaerobic conditions; therefore, after about 18 hours it needs to start inducing the gene once again.
YEN1 Expression
Since YEN1 had not yet been annotated, DNA microarrays and clustering can help narrow down the molecular function and biological process of the gene. Unfortunately, a search through the following microarrays provided little data to help support one set function for YEN1. I have tried to show below how to try to deduce an unknown from some know genes, but many of the genes which YEN1 clustered around are also unknown.
Expression of YEN1 in the PHO pathway
The PHO pathway is the regulation of genetic expression seen when there is a low concentration of phosphate in the environment. The pathway includes these topics:
1. Signal transduction mediated by Cyclin-CDK system
2. Nuclear localization of the transcriptional activator
3. Chromatin remodeling on the promoters associated with transcriptional regulation
4. Metabolic pathway of phosphate including inorganic polyphosphate
( http://cmgm.stanford.edu/pbrown/phosphate/pho_path.html)
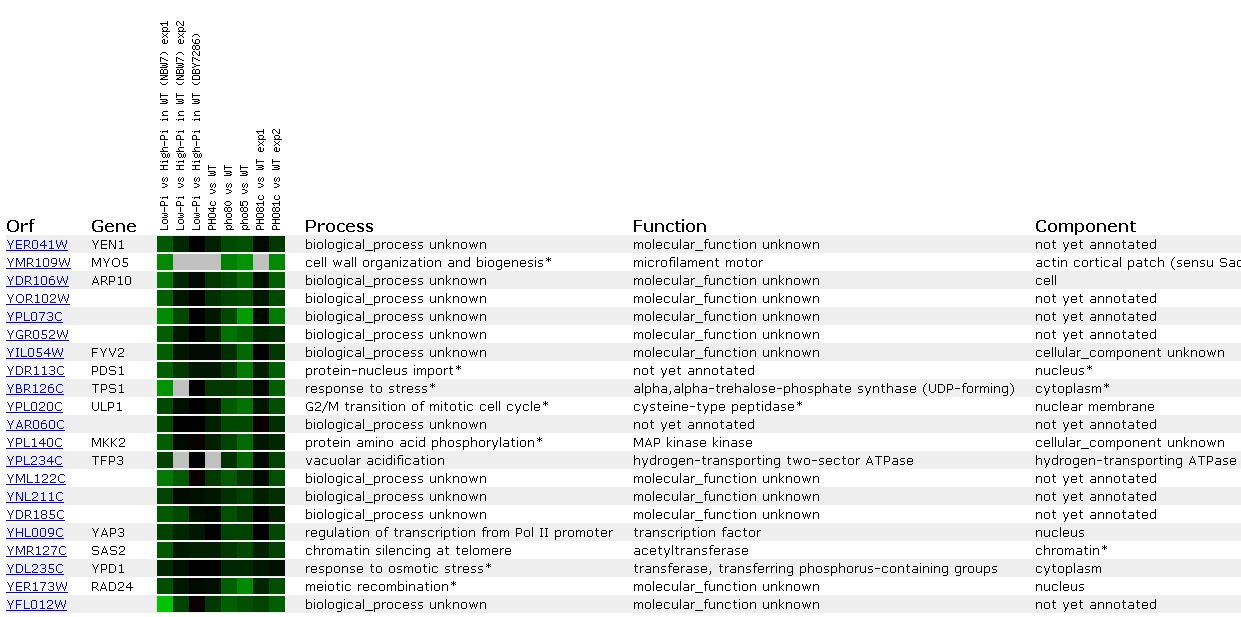
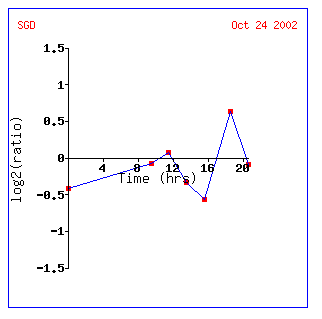
As seen above, YEN1seems to correlate well with MYO5; however, there is missing
data for four of the eight slots...not promising. There seem to be diverse
functions for all the genes listed. However, many of the genes listed are
-ase's of some kind: synthase, peptidase, ATPase, kinase...so perhaps YEN1
creates molecules by cutting them or is a signal transductor. Also, many of
the gene's proteins seem to function within the nucleus.
Expression in response to histone depletion for YEN1

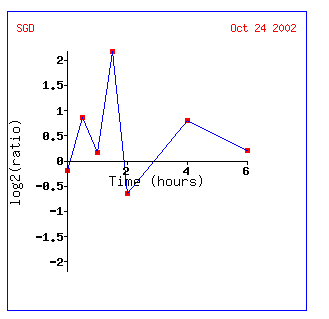
In the histone depletion microarray, I feel like YEN1 looks more similar to FUR4 which is a gene encoding a uracil transportator or ALG2 which is involved in oligosaccharide-lipid intermediate assembly. The graph of the induction then repression of this gene looks very much like that of Gln3. It supports the idea that YEN1 has a role in DNA housekeeping or genetic control (also because histones play a role in genetic control)
Expression during sporulation for YEN1
Diploid yeast cells produce haploid cells during sporulation and undergo meiosis and morphogenesis.
. 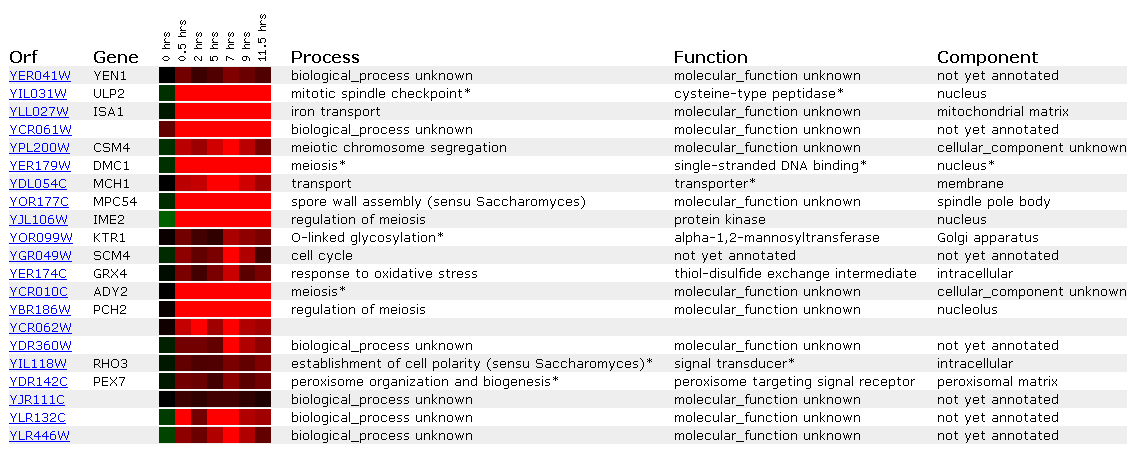
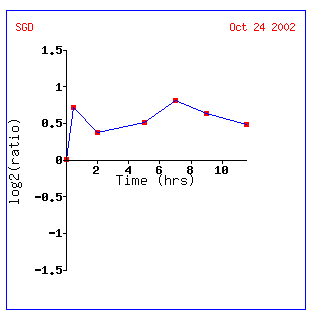
YEN1 is induced almost .8-fold the whole time and looks most like the signal transducer RHO3 and the peroxisome signal receptor PEX7. The fact that YEN1 is induced during sporulation still makes me think it encodes a nuclear protein involved in the cell cycle.
Overall, sporulation, histones, and the PHO pathway have had some genes which were highly involved in cell cycle pathways. Though I think YEN1 is in the nucleus, it may be a cell structural element. Structural genes show up a lot in the expression profiles. Mitotic spindle checkpoints, actin filament organization, meiosis regulators, G2/M transition (perhaps involving microtubules) all appear in each microarray. Thus YEN1 perhaps plays a role in cellular structure and mitosis and meiosis.
Questions or comments? Email me: laquillian@davidson.edu
© Copyright 2003 Department of Biology, Davidson College, Davidson, NC 28035