*This web page was created as an assignment for an undergraduate course at Davidson College*
LEE1 Protein information: LEE1 is an unannotated yeast gene. In my second web assignment I searched through several databases to determine information about the function of LEE1 based on its nucleotide sequence (web assignment 2). Through this search I hypothesized that LEE1 encoded a zinc finger protein. In my third web assignment I examined the expression patterns of LEE1 (web assignment 3). From the data collected in this search I hypothesized that LEE1 encoded a zinc finger protein that was involved in the regulation of transcription. In this web assignment I preformed a database search in order to investigate the functions of the LEE1 protein as well as its interactions with other proteins in a cell.
MIPS database: At the MIPs database I found a limited amount of information for the LEE1 protein. This database showed no hits for protein protein interactions or protein complex information. Figure 1 below shows the molecular weight and isoelectric point of LEE1. The figure also indicates that the MIPs database finds the LEE1 protein to have similarity to a protein found in S. pombe, homo sapiens and drosophila malongaster.
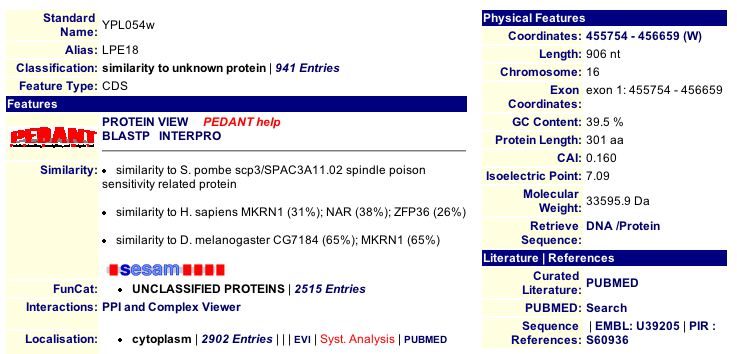
Figure 1: MIPs information regarding the unannotated yeast gene LEE1. This figure shows a molecular weight of 33595.9 Da and an isoelectric point of 7.09. The LEE1 protein has similarity to S. pombe scp3/SPAC3A11.02, H. sapiens MKRN1 and ZFP36 and D.melongaster. The amount of similarity shown in percentages next to the H. sapien and D. melongaster indicates that the sequences are not highly similar.(http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YPL054w).
SwissProt: In this database I searched for LEE1. Here I found information on the LEE1 protein. The image below demonstrates that the swiss prot database lists the molecular function of the LEE1 protein as nucleic acid binding. The database says this is based on electronic annotation. Figure 3 shows an image of a 2D gel. The red box indicates the region in which LEE1 would be found based on its molecular weight and isoelectric point.

Figure 2: Image taken from swissprot database. This website lists the molecular function of LEE1 as nucleic acid binding. The database also provides links to further information on the LEE1 protein.(http://us.expasy.org/cgi-bin/niceprot.pl?Q06701)
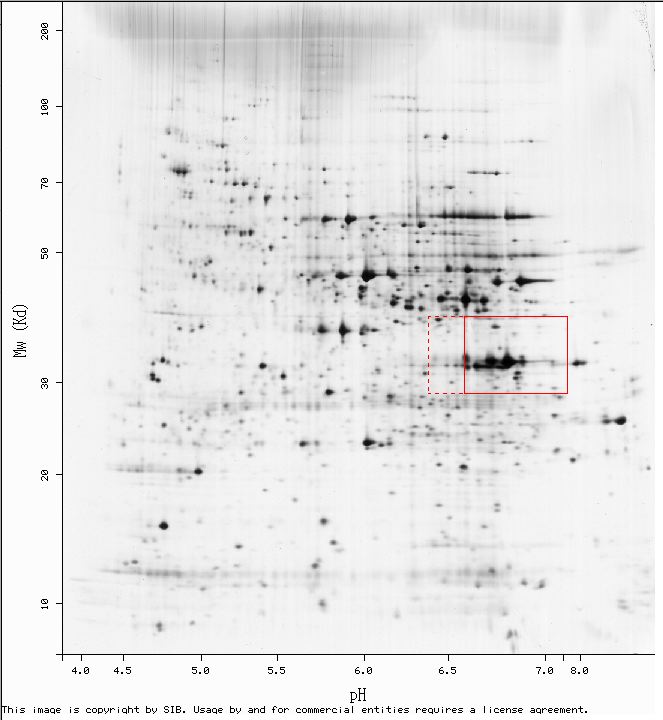
Figure 3: This image is taken from a swissprot link to expasy 2D. The red box indicates the region to which the LEE1 protein should migrate based on its molecular weight and isoelectric point. (http://us.expasy.org/cgi-bin/ch2d-compute-map?YEAST,Q06701)
Databases Searched:
PDB: When using the ID LEE1 or YPL054W I found no hits on this website.
DIP: When I searched the DIP website for LEE1 I got a hit for LEE1. However, no graph was made because the DIP website does not list LEE1 as interacting with any other proteins.
Benno Figure 1 pdf. When searching this integrated protein circuit diagram I found no hits for LEE1.
The Y2H database showed no hits for LEE1.
Conclusions:
Throughout my investigation of my favorite uannotate yeast gene, LEE1, I have come to the conclusion that its protein functions in RNA transcription or some form of DNA binding and regulation. Although I did not find much information for the LEE1 protein, the information I found supported my hypothesis. The SwissProt database listed LEE1 as having a molecular function of nucleic acid binding. The website also indicates that this inference was made based on electronic annotation. Therefore, the SwissProt database came to a similar conclusion when annotating the LEE1 gene through electronic database searches, just as I have done. The more tests that are performed on LEE1 the more information we will have on this protein. A greater amount of information would be need in order to further test my hypothesis.
Possible Further Investigations:
The LEE1 protein has many areas in which it could be further investigated. To begin an investigation the protein's localization could be examined. I hypothesized that this protein is involved in regulation of transcription or is some form of DNA binding protein. Therefore I would expect the protein to localize to the nucleus. In order to carry out this investigation I would create fluorescent label an antibody for the LEE1 protein. I would then place that antibody in a yeast cell and see where the fluorescence appeared.
After localization a yeast 2 hybrid experiment could be used to further identify protein protein interactions. In this experiment the LEE1 protein is used as a bait which is fused to the DNA binding domain of a transcription factor. Possible prey proteins, which might include known DNA binding proteins or transcription factors, are then used as prey and are fused to the activation domain of the same transcription factor. If the bait and prey proteins interact then the transcription factor will initiate transcription of some identifiable gene (Campbell, 2002). This experiment would allow me to further test my hypothesis. I would expect the LEE1 protein to interact with other transcription factors.
In order to further test my hypothesis more DNA microarray experiments could be conducted. I would use a DNA microarray of LEE1 during the cell cycle to test my hypothesis. I would expect the induction or repression of LEE1 to be greatly altered during DNA replication or during cellular growth prior to division. These changes in LEE1 transcription would support my hypothesis that LEE1 functions as a transcription factor or as a DNA binding protein.
References:
Campbell, M., Heyer L. Discovering Genomics Proteomics & Bioinformatics. Cold Spring Harbor Laboratory and Benjamin Cummings. San Francisco. 2003
Schwikowski, Benno, Uetz P., Fields S., A network of protein-protein interactions in yeast. Nature Biotechnology. 18: 1257-1261.