MIF2 is an essential gene that encodes for a centromeric protein involved in mitotic spindle structural assembly and DNA binding at the centromere. It is located on chromosome eleven at cM position -43. The coding sequence for MIF2 is:
ATGGATTATATGAAATTGGGGCTTAAGTCCCGTAAAACTGGTATTGATGTTAAGCAAGATA TACCCAAAGATGAATACAGCATGGAAAATATTGATGATTTTTTCAAAGATGATGAAACT
AGTCTTATCAGTATGAGAAGGAAAAGCAGAAGAAAATCATCGCTTTTCTTACCATCAACG
TTAAATGGCGATACTAAGAACGTATTACCGCCATTTCTACAGTCATATAAATCTCAAGAT
GATGAAGTTGTCCAAAGCCCATCTGGGAAAGGCGATGGATCAAGACGATCATCTTTGTTA
AGCCATCAATCAAACTTCCTGAGTCCAGCCAATGATTTCGAGCCTATTGAGGAAGAACCA
GAACAAGAAGAAAATGATATCAGAGGCAATGATTTTGCCACACCAATCACACAGAAATTG
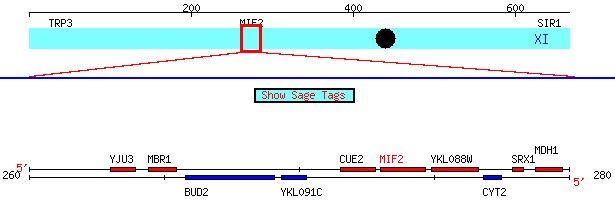
Gene Sequence for a region of MIF2/YKL089W

Figure 1. Screen capture depicting the chromosomal location of MIF2 and its neighboring ORFs.
Red depicts the Watson strand, while blue depicts the Crick strand. ORF Map
MDYMKLGLKSRKTGIDVKQDIPKDEYSMENIDDFFKDDETSLISMRRKSRRKSSLFLPST
LNGDTKNVLPPFLQSYKSQDDEVVQSPSGKGDGSRRSSLLSHQSNFLSPANDFEPIEEEP
EQEENDIRGNDFATPITQKLSKPTYKRKYSTRYSLDTSESPSVRLTPDRITNKNVYSDVP
DLVADEDDDDRVNTSLNTSDNALLEDELEDDGFIPESEEDGDYIESDSSLDSGSDSASDS
DGDNTYQEVEEEAEVNTNDNEDDYIRRQASDVVRTDSIIDRNGLRKSTRVKVAPLQYWRN
EKIVYKRKSNKPVLDIDKIVTYDESEDEEEILAAQRRKKQKKKPTPTRPYNYVPTGRPRG
RPKKDPNAKENLIPEDPNEDIIERIESGGIENGEWLKHGILEANVKISDTKEETKDEIIA
FAPNLSQTEQVKDTKDENFALEIMFDKHKEYFASGILKLPAISGQKKLSNSFRTYITFHV
IQGIVEVTVCKNKFLSVKGSTFQIPAFNEYAIANRGNDEAKMFFVQVTVSEDANDDNDKE
LDSTFDTFG
MIF2 encoded protein sequence made up of 549 amino acids
Molecular Function
Mif2p, the protein encoded by the yeast gene MIF2, functions in DNA binding at the centromere. MIF2 protein was first determined to play a role in the binding of DNA at the centromere based upon the protein's interactions with S. cerevisiae centromere proteins and by in vivo crosslinking to centromeric DNA. Confirmation of MIF2 protein localization and functionality at the centromere was determined using a technique referred to as a GST pull down assay. A GST pull down assay is a technique used to determine whether two proteins interact with one another physically (Hemmerich, 2000). The confirmation of MIF2 protein interaction with a number of known centromeric proteins, such as Cbf1p and CBF3, eventually led to the discovery that in fact MIF2 protein also functions in centromeric DNA binding through the connections it establishes among other essential centromeric proteins, which ultimately results in structural assembly and maintainence at the centromere.
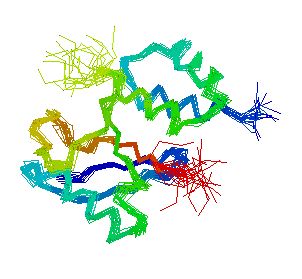
Figure 2. Screen capture of model of MIF2 encoded protein
Biological Process
MIF2 is responsible for encoding a protein essential to the mitotic process. The function of the MIF2 encoded protein was determined by generating mutants that lacked the functional protein, which in essence eliminated the protein. The lack of MIF2 encoded protein resulted in the inability of the chromosomes to undergo normal segregation during mitosis due to improper spindle orientation and structural assembly. Mutant MIF2 studies led to the determination that Mif2p, the protein encoded by MIF2, was required in order to maintain the structure of the spindle prior to anaphase spindle elongation (Brown, 1993). Prior to the elongation step, mutant mif2p spindles were observed to halt the mitotic processs and undergo both a loss of spindle polarization and structural stability. As a result, MIF2 was established to be responsible for encoding a protein that plays a crucial role in mitotic spindle assembly and maintainence.
Cellular Component
Based upon its interactions with a multitude of centromeric proteins at a particular site, SGD lists the MIF2 protein as being located at the specialized centromeric nucleosome (Hyland 1999). The specialized centromeric nucleosome is the core region around which centromere DNA is wrapped.
__________________________________________________________________________________________________________________________________________________________
YKL091C is an ORF located on chromsome eleven in relatively close proximity to MIF2
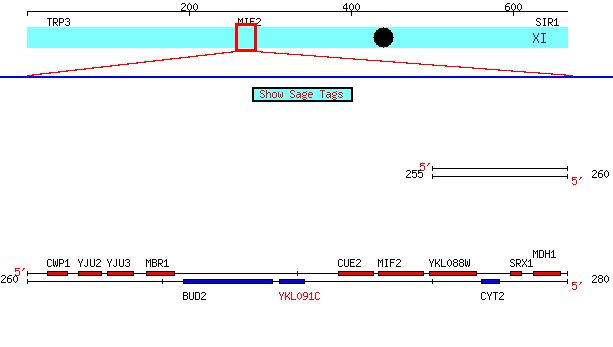
Figure 3. Screen capture depicting the location of the YKL091C ORF
YKL091C is located on chromosome eleven, near the genes BUD2 and CYT2.
Using the YKL091C gene sequence in GCG sequence format, I performed a BLASTn search in order to determine the sequence which have significant alignments with the sequence in question. The results of the BLASTn search are listed on the table below:
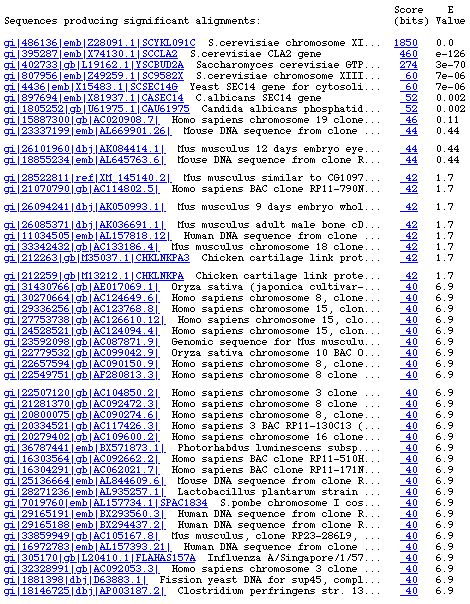
Figure 4. Screen capture of BLASTn results
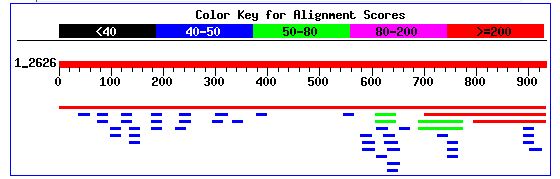
Figure 5. Distribution of BLASTn hits for the YKL091C sequence
While the BLASTn results did not yield many significant sequence alignments, a BLASTp of the protein sequence resulted in a number of hits, which corresponded to the YKL091C protein sequenece to a high degree. The distribution of the BLASTp hits is shown below:
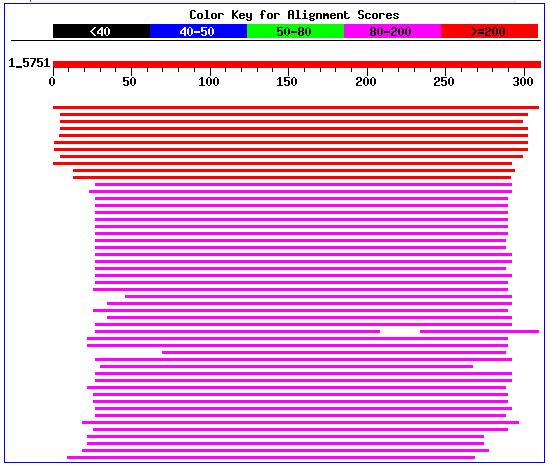
Figure 6. Screen capture of alignments from BLASTp search
In order to evaluate the possible function of the protein encoded by the gene YKL091C, I analyzed those proteins, with the highest degree of alignment with the query sequence. Similar proteins from both a yeast (S. cerevisiae) and non-yeast (Lotus japonicus) species were analyzed on a functional basis. The related protein from the Lotus japonicus acts as a phosphatidylinositol transfer-like protein (PITPs), which modulates signal transduction pathways and membrane trafficking functions in eukaryotes. The protein also exists as PITP Sec14p in S. cerevisiae, which is a cystolic factor involved in protein movement from the Golgi complex. From the evidence concerning the of these related proteins, I conclude that the gene YKL091C encodes for a PITP.
___________________________________________________________________________________________________________________________
References
"MIF2/YKL089W." Saccharomyces Genome Database. Last Update: 2003. <http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?sgdid=S0001572>
"YKL091C." Saccharomyces Genome Database. Last Update: 2003. <http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=YKL091C>
Hemmerich P, et al. Interaction of yeast kinetochore proteins with centromere-protein/transcription factor Cbf1. Proc Natl Acad Sci U S A 97(23):12583-8 (2000).
Brown MT, et al. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J Cell Biol 123(2):387-403 (1993).
Hyland KM, et al. Ctf19p: A novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol 145(1):15-28 (1999).
"Protein Explorer." <http://www.pdb.org>