Using Public Databases for DNA Microarrays to Study My
Annotated and Non-annotated Genes
In this page, I have searched
for other genes that have similar expression profiles to my genes in different
experiments.
Using the idea of guilt by association, I looked for clustering patterns of
my gene with other genes of known functions,
and how my gene was regulated the same way as other genes of known functions,
so that I could infer the function of my gene better.
My Annotated Gene: SSD1 (YDR293C)
FUNCTION
JUNCTION
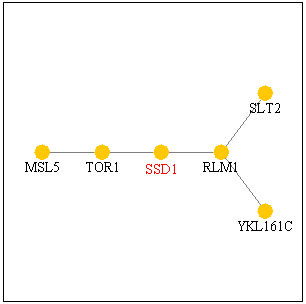
Figure 1. SSD1 protein interacts
with TOR1p and RLM1p as shown in this pathway. Image from SGD
Function Junction.
SGD provided very interesting information on TOR1 as I connect this gene to
my SSD1 gene.
I have found for my last web page that the molecular
function of SSD1 was RNA binding and modification,
and the biological process was cell wall organization and biogenesis, and the
cellular component was the cytoplasm.
Additionally, I found out that SSD1 was implicated in the control
of the cell cycle G1 phase.
SGD documents that TOR1 has
the molecular function of phosphatidylinositol 3-kinase activity and protein
binding,
and the biological process was involved in G1 phase of mitotic cell cycle,
meiosis, regulation of cell cycle,
ribosome biogenesis, and signal transduction. Also, TOR1 has the
cellular component of Golgi membrane,
endosome membrane, plasma membrane and vacuolar membrane. This information
on TOR1 may
shed light on
the SSD1 protein's role in cell cycle G1 phase, and the cellular component
of TOR1
may suggest
specific locations where SSD1 may be found within the cytoplasm.
Information from SGD on RLM1 also
had significant connection to my SSD1 gene.
Molecular function of RLM1 is DNA bending activity, DNA binding and
transcriptional activator activity,
and the biological process is cell wall organization and biogenesis, which
is similar to the biological process of SSD1,
and
positive
regulation of transcription from Pol II promoter and
signal transduction.
The cellular component of RLM1 is the nucleus.
SLT2,
connected to RLM1 in the pathway, also participates in cell wall organization
and biogenesis,
which is
similar
to the biological process to SSD1, and the cellular component of SLT2 includes
cytoplasm,
where SSD1 gene products are found.
Global gene expression during short-term ethanol stress in Saccharomyces
cerevisiae,
Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B., June 2001.
FEBS
Lett. 2001 Jun 1;498(1):98-103.
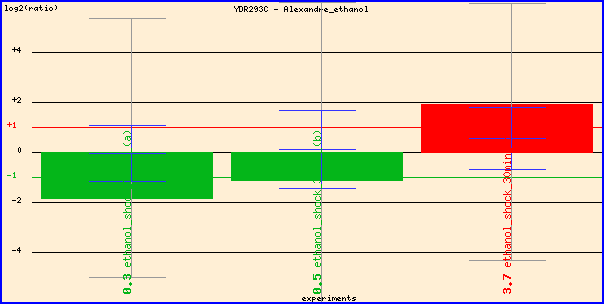
Figure 2. Expression of SSD1 during ethanol stress. The researchers reversed
the dye color of experimental
and control conditions in the last
red
column, so the red column actually shows repression of SSD1 during ethanol stress.
It makes sense that SSD1, which participates in cell wall organization and biogenesis,
is repressed in the presence of ethanol,
so yeast can induce environmental stress response and turn on energy metabolism
genes better.
YEAST CELL CYCLE ANALYSIS
PROJECT
This site complements the paper published in 1998 by Paul T. Spellman,
Gavin Sherlock, Michael Q. Zhang,
Vishwanath R. Iyer, Kirk Anders, Michael B. Eisen, Patrick O. Brown, David
Botstein, and Bruce
Futcher.
The researchers set out to identify all genes regulated by the yeast Saccharomyces
cerevisiae cell cycle using microarrays.
The full text is available here.
Since my gene SSD1 showed pathway relevance to TOR1,
heavily involved in cell cycle,
I used this database to analyze my gene.
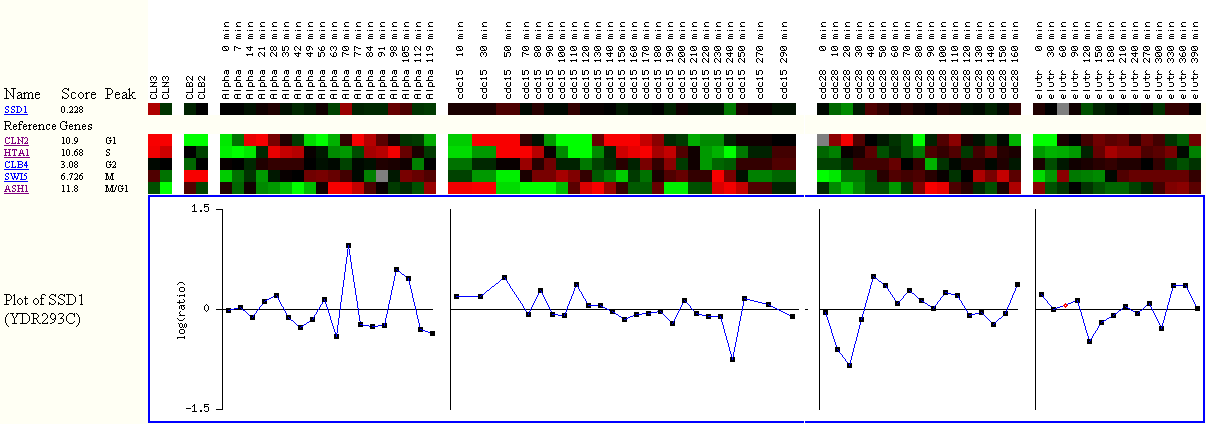
Figure
3. The Score, algorithmically generated, shows how well the gene is cell cycle
regulated.
The higher the score, the better the expression data indicated that
the gene is cell cycle regulated.
The Peak shows where the gene was expressed the most during the cell cycle.
The red color means that the gene was induced, while green
means repressed,
and black
means almost no change in gene expression between experiment and control.
The reference genes represent each cell cycle. For example, CLN2 represents the
cluster of genes
that were induced the most during the G1 phase.
I observe slight induction in SSD1 expression during the alpha factor arrest
experiment,
which corresponds to G1 phase of the cell cycle. However, the overall score
for SSD1 is 0.228,
which is relatively small, compared to 10.9 of CLN2. The small score, according
to the researchers,
suggests weak link between SSD1 expression profile and other genes that are
very cell cycle regulated.
This figure reaffirms why gene ontology does not list cell cycle regulation
as the biological process of SSD1.
EXPRESSION
CONNECTION
Expression during the cell cycle,
Stanford University, Cold Spring Harbor
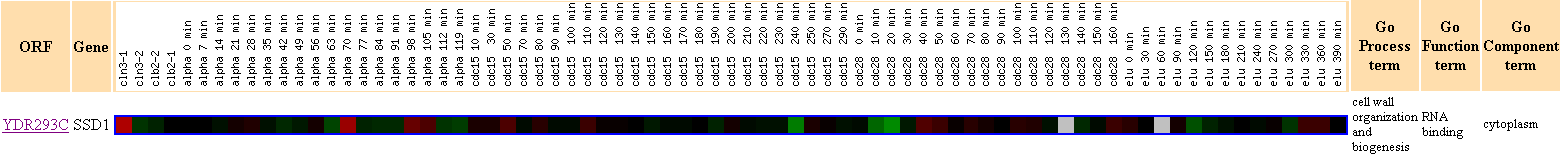
Figure 4. This figure shows that there were no other genes with Pearson
correlation >0.8 to the expression profile of SSD1.
This finding reaffirms what I have suggested earlier in Figure 3,
that SSD1
does not seem to have as strong connection to the cell cycle regulation as
I have expected earlier.
Expression during the diauxic
shift, Stanford University
Since the diauxic shift of yeast is involved with going from oxygen
rich
environment
to no oxygen,
the yeast cells would have to focus on energy metabolism and stress response,
rather than cell wall organization or biogensis,
so I assume that expression of SSD1 would be repressed during diauxic shift,
and would be clustered with other repressed genes.
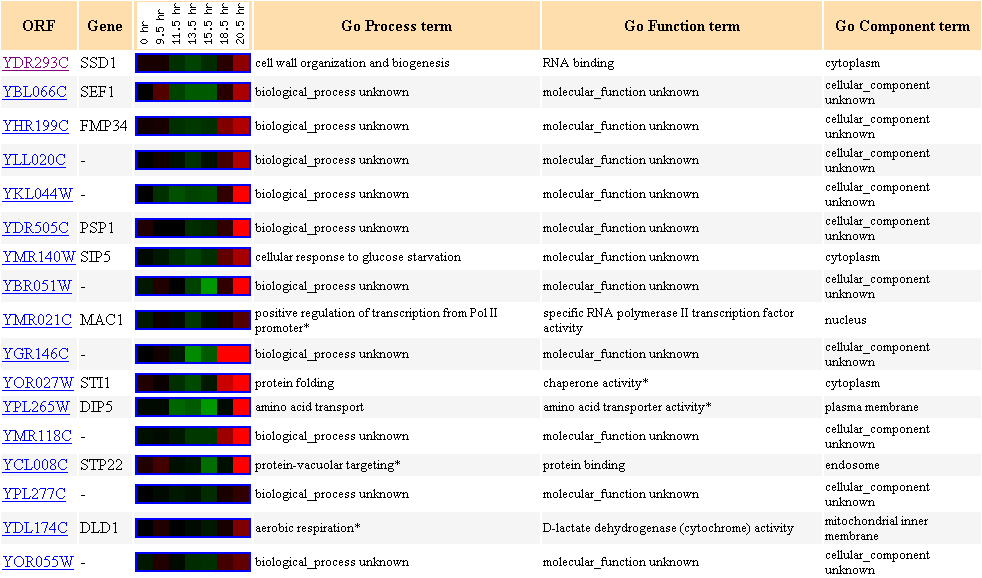

Figure 5. Cluster of
genes with similar expression file to SSD1 during diauxic shift in yeast.
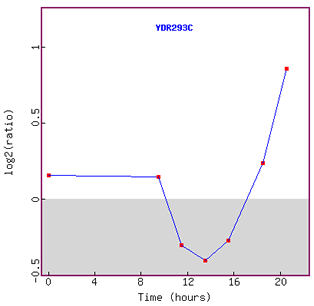
Figure 6. Graph of SSD1 expression during diauxic shift over time.
As I have predicted eariler, expression of SSD1 was repressed initially.
However, interestingly enough, after about 13 hours, expression of SSD1 was induced
again.
I assume that yeast was able to adjust to the new anaerobic environment in about
13 hours metabolically,
so started to turn on other functional genes such as SSD1.
Expression in response to alpha-factor
(over time), Rosetta Inpharmatics
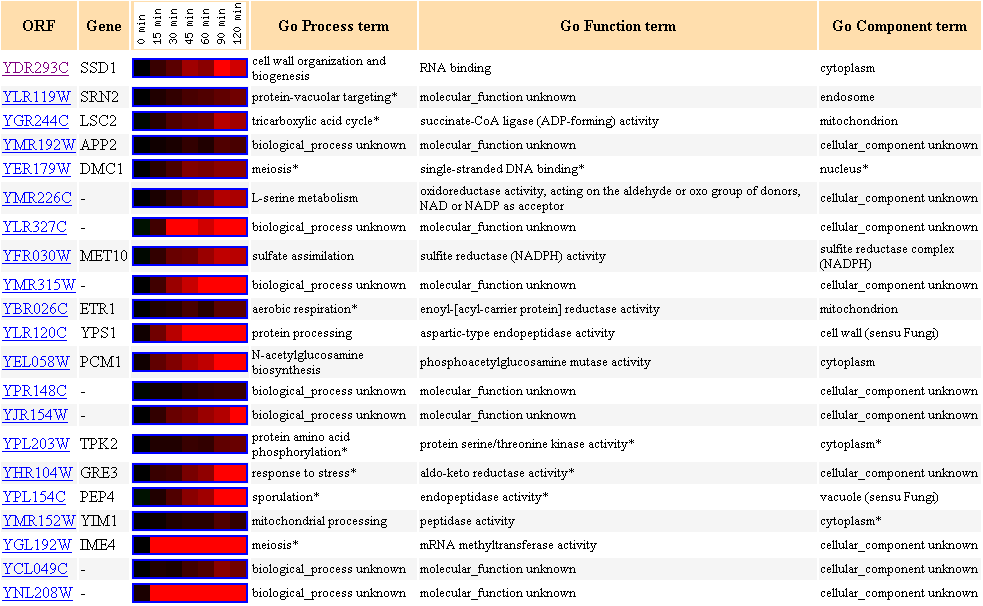
Figure 7. Cluster of genes with similar expression profiles in response to alpha-factor
over time with SSD1 as the clustering seed.
Alpha-factor mating pheromone arrests yeast cell cycle in G1 (Zymo
Research,
200l).
There are two genes (DMC1 and IME4) that are involved in meiosis in this cluster.
Other biological processes of genes in this cluster include protein processing,
mitochondrial
processing,
TCA cycle and aerobic respiration.
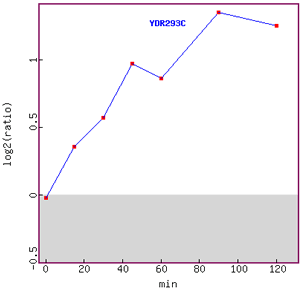
Figure 8. Graph of SSD1 expression over time in response to alpha-factor.
Induction of expression is clearly shown in this figure, although the graph tails
off at 120 minutes.
Since SSD1 has been associated with the cell cycle in G1 phase,
I can hypothesis that the gene was induced more as the cell cycle was
arrested
in
G1
phase.
Expression in response to alpha-factor
(various concentrations), Rosetta Inpharmatics
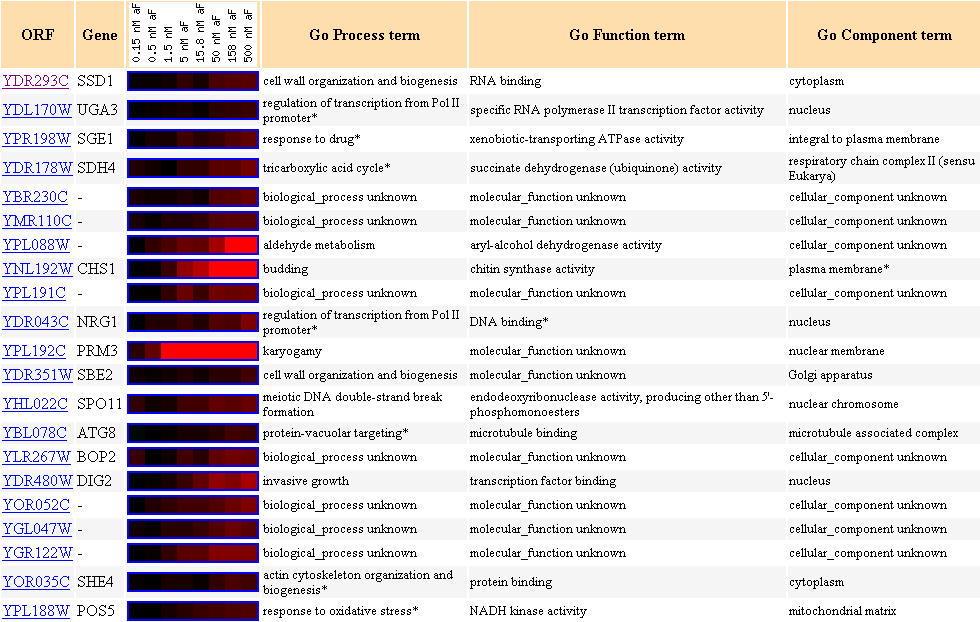
Figure 9. Cluster of genes with similar expression profiles in response to different
concentrations of alpha-factor
with SSD1 as the clustering seed.
Another gene (SBE2) with biological process of cell wall organization and biogenesis
is clustered here.
NRG1 participates in regulation of trascription from Pol II promoter, and its
molecular function is DNA binding,
and its cellular component is the nucleus, and these parameters are almost identical
to RLM1,
which I have discussed
earlier in Figure 1. Other notable biological processes of genes included
in this cluster are regulation of transcription,
and actin cytoskeleton organization.
Expression during sporulation,
UCSF, Stanford University
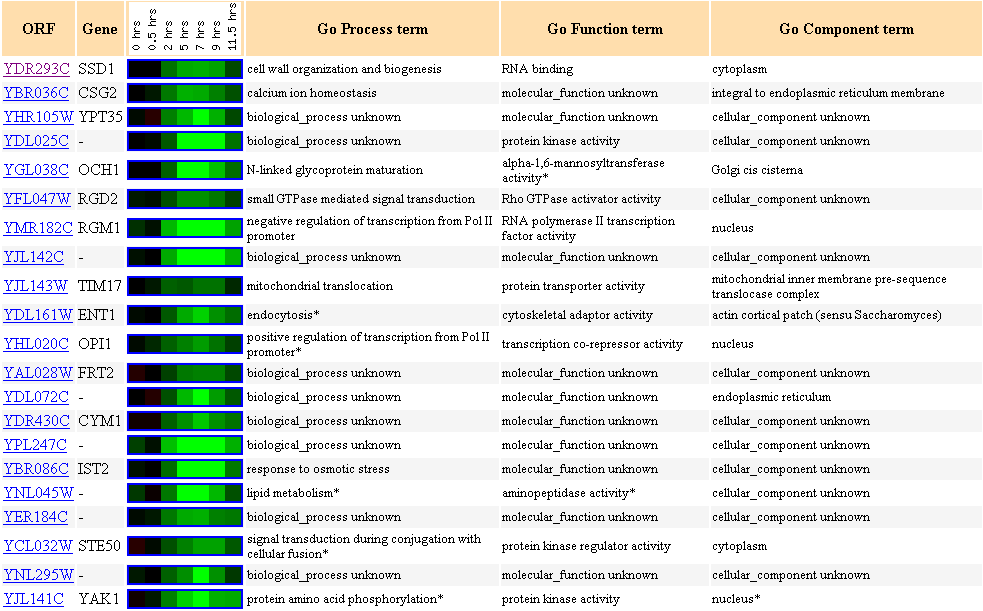
Figure 10. Cluster of genes with
similar expression profiles during sporulation with SSD1 as the clustering seed.
There are many genes with unknown biological process and molecular function
included in this cluster.
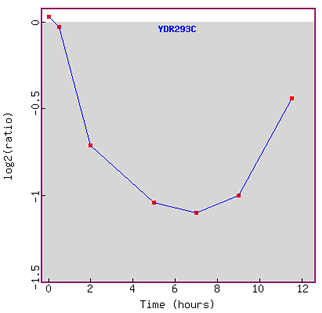
Figure 11. Graph of SSD1 expression
profile during sporulation over time.
Initially SSD1 is repressed, but is induced as time went on.
Since yeast produces spores through meiotic divisions, I am surprised to observe
here that SSD1 is initially repressed,
because I would assume that cell wall organization should be activated even
more while sporulation occurs.
What I observe here is a very interesting trend of significant initial repression
followed by induction as time went on.
I can understand why lipid metabolizing gene would be repressed during sporulation
(YER184C),
because it would be more important and energy efficient for yeast to focus on
producing spores than metabolizing lipid.
The Bottom Line about SSD1
Previously, I have found that SSD1(YDR293C) has biological process
of cell wall organization and biogenesis,
and molecular function of RNA binding and modification, and cellular component
of cytoplasm.
The microarray data I have presented above reinstate these findings. For example,
SSD1 does not participate in
yeast metabolic activities or stress response significantly, because I have
found that the gene becomes repressed under
stress conditions, such as exposure to ethanol and during diauxic shift up to
13 hours. Also, my findings reaffirm
why gene ontology does not include "cell cycle regulation" or "cell
growth regulation" as the biological process,
because even though much evidence suggests that SSD1p interacts with other proteins
heavily involved in cell cycle regulation,
the expression profile of SSD1 during cell cycle shows weak link to expression
profiles of other yeast cell cycle regulation genes.
SSD1 expression profile clearly does not cluster with other genes that show
distinctive expression profiles synchronized
with the yeast cell cycle. I hypothesize that SSD1p plays a significant role
in the cell cycle pathway, but does not
participate in cell cycle or growth regulation directly. Another hypothesis
I have considered while performing this search was that
SSD1 may be involved in regulation of transcription. I see that SSD1p interacts
directly with BLM1, whose molecular function is
DNA bending activity, DNA binding and transcriptional activator activity, and
the biological process is cell wall organization
and biogenesis, which is similar to the biological process of SSD1, and positive
regulation of transcription from Pol II promoter
and signal transduction. Also, in Figures 5 and 9, SSD1 showed similar expression
profiles to MAC1 and NRG1.
MAC1's biological process is positive regulation of transcription from Pol II
promoter, and the molecular function is
specific RNA polymerase II transcription factor activity. NRG1's biological
process is regulation of transcription from Pol II promoter,
and its molecular function is DNA binding.
My Non-Annotated Gene: YDR288W
FUNCTION
JUNCTION

Figure 12. Disappointingly, Function Junction does not list any
other genes interacting with YDR288W.
It does not mean YDR288Wp does not interact with other proteins; the database
is growing all the time.
EXPRESSION
CONNECTION
Expression during the cell cycle, Stanford University, Cold Spring Harbor

Figure 12. Expression profile of
YDR288W during the cell cycle.
I expected to see more genes clustered in this figure, because I hypothesized
that YDR288W had something to do with the cell cycle,
given the gene's homology to the human MAGE (melanoma
antigen-encoding gene) family.
Expression in response to alpha-factor (over time), Rosetta Inpharmatics
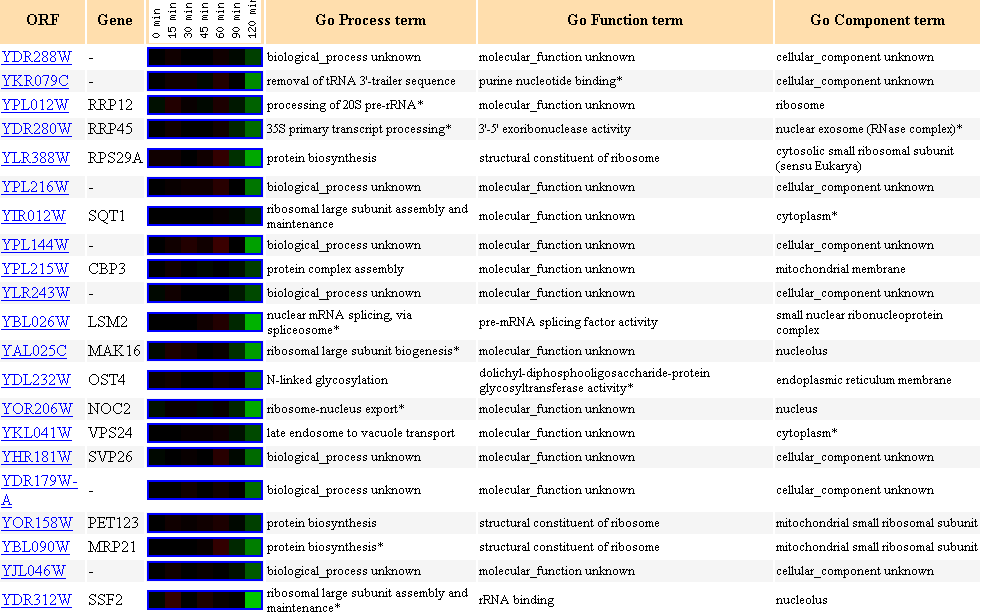
Figure 14. Cluster of genes showing
similar expression profiles to YDR288W during exposure to alpha-factor over
time.
I have suggested in my last page that YDR288W possibily
participates in cell growth or cell cycle regulation,
because YDR288W showed homology to the human MAGE (melanoma antigen-encoding
gene) family.
As suspected, I see that YDR288W expression profile clustered with many other
genes involved in cell cycle G1,
because alpha-factor arrests the cell cycle in G1 as I have referenced earlier.
Notable biological processes of other genes in this cluster are ribosomal large
subunit assembly,
protein biosynthesis. Also note CBP3, whose biological process is protein assembly,
found in the ribosomal membrane,
because I have suggested previously that YDR288W may be a integral membrane
protein according to the Kyte-Doolittle hydropathy plot.
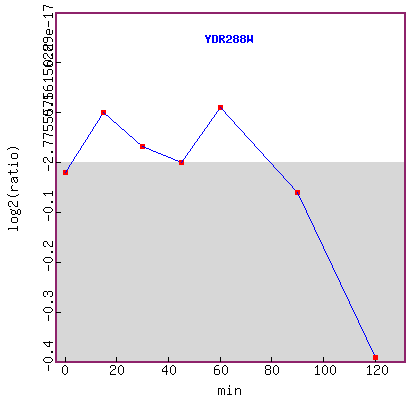
Figure 15. Graph of YDR288W expression
during exposure to alpha-factor over time.
It seems that YDR288W expression level did not have a distinctive change trend
up to 60 minutes,
but expression was repressed after 60 minutes.
Expression in response to alpha-factor (various concentrations),
Rosetta Inpharmatics
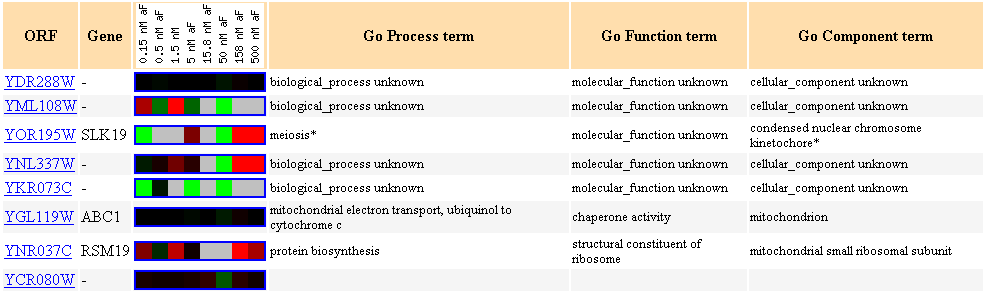
Figure 16. Cluster of genes showing similar expression profiles to YDR288W during
exposure to different concentraions of alpha-factor.
It is interesting to see that protein biosynthesis came up again, and that two
mitochondrial genes surfaced again,
just as there were other mitochondrial genes included in Figure 15.
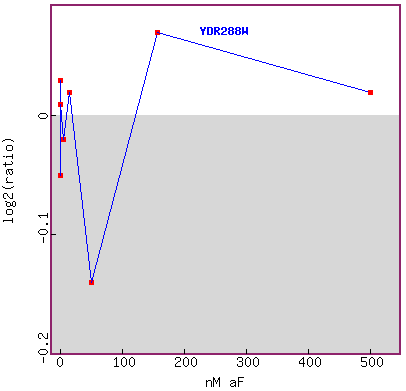
Figure 17. Graph of YDR288W expression during exposure to different concentrations
of alpha-factor.
The researchers tested many low concentrations, and skipped many concentrations
in between up to about 1000 fold.
I don't think this graph tells me anything definitive in YDR288W expression
trend during exposure to different
concentrations of alpha factor. I would like to see more data points in this
figure.
Expression in response to environmental changes, Stanford
University
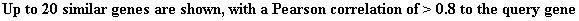
Figure 18. YDR196C
clustered with YDR288W during environmental stress and changes. Biological
process, molecular function and cellular component of YDR196C is given to the
right of the figure.
Strong induction of both genes is observed during nitrogen depletion.
Expression during the diauxic shift, Stanford University
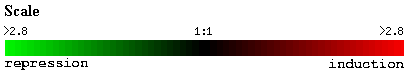
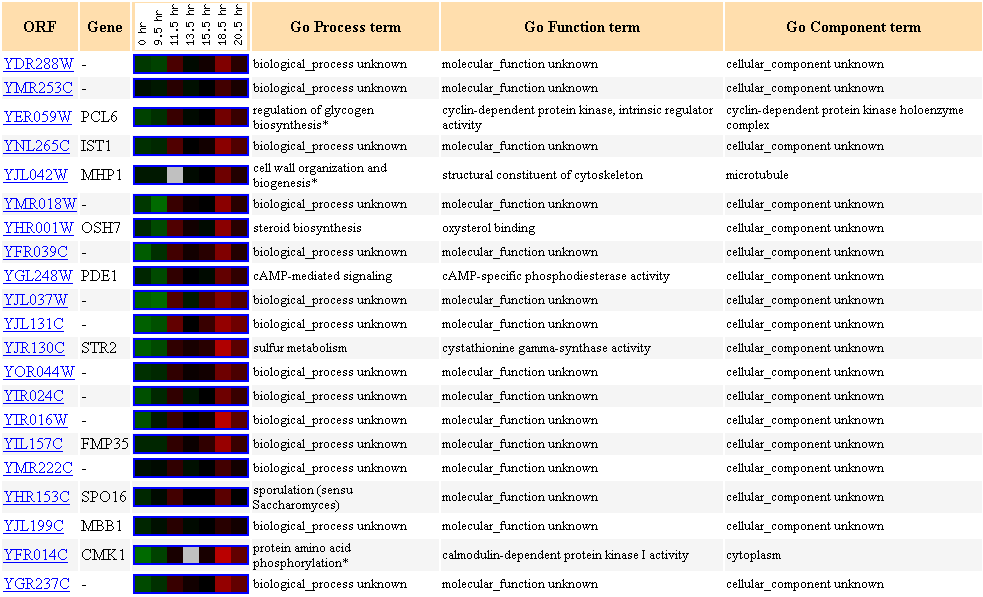
Figure 19. Cluster of genes showing
similar expression profiles to YDR288W during the diauxic shift.
Many genes in this cluster are not characterized. Two different types of kinase
are clustered here (CMK1 and PCL6).
Since YDR288W is possibly an integral membrane protein, I had hypothesized earlier
that YDR288W
may be post-translationally modified, such as being phosphorylated (Link
to my last page).
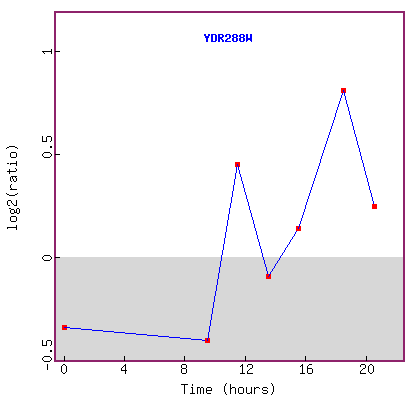
Figure 20. Graph of YDR288W expression during diauxic shift.
This figure seems to be inconclusive in explaining the expression trend of the
gene,
because the lowest log2 ratio of repression is about -0.5 fold,
and the highest is about +0.8 fold, and in 20 hours, the log2 expression ratio
is back down to about +0.3.
If this experiment was replicated, it would be helpful to see error bars on
each point.
Expression in response to DNA-damaging agents, Stanford
University

Figure 21. YDR288W clustered with YDR196C in response to DNA-damage.
Note that YDR196C clustered with YDR288W before in Figure 18, during environmental
changes.
Also note that YDR196C has the molecular function of dephospho-CoA kinase activity,
which may suggest that YDR288W may also be involved in kinase activity.
Expression in response to histone depletion, Stanford
University
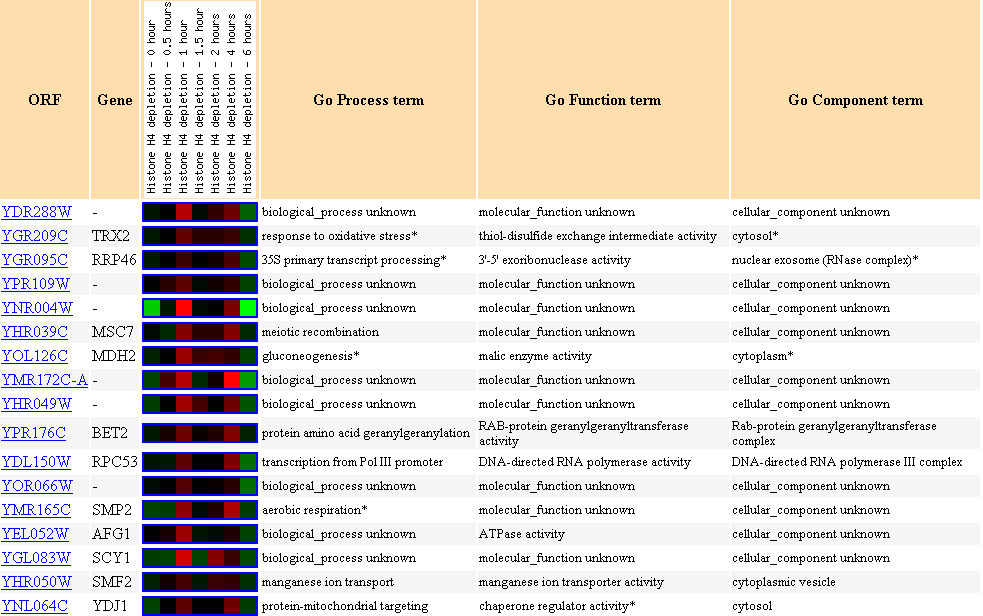

Figure 22. Cluster of genes showing
similar expression profiles to YDL288W in response to histone deletion.
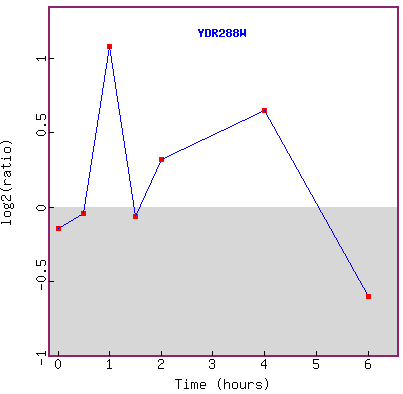
Figure 23. Graph of YDR288W expression
in response to histone deletion over time.
YEAST
CELL CYCLE ANALYSIS PROJECT

Figure 24. Expression of YDR288W
during the cell cycle with reference genes.
The score for YDR288W is 0.447, which is higher than 0.228 of my annotated gene
SSD1,
suggesting that YDR288W is more cell cycle regulated compared to SSD1.
But there has been evidence that SSD1 is implicated in the yeast cell cycle
G1,
and we observe slight induction of SSD1 during G1 in Figure 3.
So, the fact that YDR288W has a higher score than SSD1 is interesting,
because it implies that YDR288W may potentially be linked to the cell cycle
in significant ways
that we just don't know about yet.
The Bottom
Line about YDR288W
My non-annotated gene YDR288W expression profile clustered with YDR196C twice
in the DNA damge experiment
and the environmental changes experiment. Both genes are located on the yeast
chromosome IV,
only about 530 basepairs apart. YDR196C participates in dephospho-CoA kinase
activity, and I wonder
if YDR288Wp interacts with YDR196C in any physical way, or if both genes have
the same promotor.
If they turn out to have the same promotor, then YDR288W would also participate
in kinase activity,
which goes along with what I had hypothesized earlier that YDR288W may be post-translationally
modified.
Even though Figure 12 shows that YDR288W did not cluster with any other genes
during the cell cycle,
I cannot reject the idea that YDR288W is involved in the cell cycle regulation,
because evidence presented here
is in progress or generally inconclusive. Expression of YDR288W during exposure
to alpha-factor over time suggests that
YDR288W may be involved in biosynthesis or assembly of proteins and other molecules.
However, I do not find any conclusive evidence to what I have hypothesized earlier
in my last page.
I do not find sufficient evidence to conclude that YDR288W is an integral membrane
protein from the data presented above,
nor that YDR288W is within the cell cycle signalling pathway.
Sources
SGD database. 2003. Stanford University.
<http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=TOR1>
<http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=RLM1>
<http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=SSD1>
<http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=SLT2>
Expression Connection. 2003.
<http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl>
Function Junction. 2003.
<http://db.yeastgenome.org/cgi-bin/SGD/functionJunction>
Zymo Research. 2003.
<http://www.zymor.com/y1001-frame.htm>