On the last two web assignments, I studied the annotated Saccharomyces cerevisiae gene PCP1 and the non-annotated gene YGR102C. Here, I continue my investigation by exploring proteomics databases and PDF files to learn more about the proteins encoded by my favorite yeast genes. The data found on this webpage is available at the following sources:
▪ Protein Data Bank (PDB): A worldwide repository for the processing and distribution of 3-D biological macromolecular structure data.
▪ PROWL database: This database allows the user to search a wide range of species. Provides the molecular weight and isoelectric point of proteins is determined by sequence analysis.
▪ Yeast Two-hybrid: This website provides data regarding protein-protein interactions in yeast.
▪ Database of Interacting Proteins (DIP): The DIP is a useful source of information when studying protein-protein interactions. Interactions are depicted in the form of a graph where the nodes denote proteins and edges join the proteins which interact directly with each other. The following legend will be used to interpret the graphs obtained from this database:

▪ The Benno Schwikowski figures: Schwikowski, Fields and Uetz collated 2709 interactions among 2039 different proteins. They synthesized an interaction map for yeast which contains 2358 interactions among 1548 proteins in the yeast proteome.
▪ MIPS Comprehensive Yeast Genome Database (CYGD): This database provides information about the molecular structure and functional network of Saccharomyces cerevisiae.
▪ What Is There (WIT) Database: Supplies information about the function assignments made to genes and the development of metabolic models.
REVIEW
PCP1 is a well characterized gene located on chromosome VII in Saccharomyces cerevisiae. The systematic name for this gene is YGR101W. The Gene Ontology information about PCP1 is as follows:
Molecular function: peptidase activity i.e. catalysis of the hydrolysis of peptide bonds.
Biological process: mitochondrial intermembrane space protein import; mitochondrion organization and biogenesis.
Cellular component: mitochondrion (in the mitochondrial inner membrane).
PROTEOMIC ANALYSIS:
Structure:
►Protein Data Bank (PDB): A search for the structure of the PCP1 encoded protein yields no results.
►PROWL: Sequence analysis calculates the molecular mass of PCP1p as 38790.152 Da, and the isoelectric point (pI) as 10.4.
Protein Interaction Networks:
►Yeast Two-hybrid: According to this database PCP1 has no interactions with other proteins.

Figure 1: PCP1, also known as YGR101W has no interactions.
► The Database of Interacting Proteins (DIP):
The graph below depicts the protein-protein interactions for PCP1.
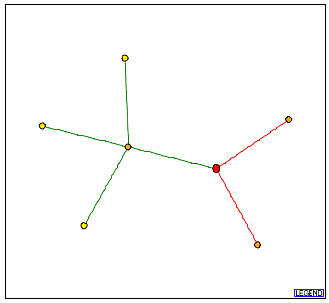
Figure 2: Graph obtained from the DIP which shows the protein-protein interactions for PCP1.
In this figure, the red node denotes the PCP1 protein. It directly interacts with three other proteins:
CCP1 protein - This is the orange node connected to PCP1p with a green line. This means that CCP1p directly interacts with PCP1p and this interaction has been verified by one or more methods. According to the Saccharomyces Genome Database (SGD), CCP1 is involved in oxidative stress response, and is located in the mitochondrion and the mitochondrial intermembrane space.
FMP12/YHL021C protein - This corresponds to a hypothetical ORF with unknown biological process and molecular function. However, this authentic, non-tagged protein was localized in the mitochondria.
YJL213W protein - This corresponds to a hypothetical ORF whose gene ontology information is as yet unknown.
► The Benno Schwikowski figures: PCP1 is not one of the proteins in the interaction map.
►MIPS: Two physical interactions are listed. The proteins with which the PCP1 encoded protein interacts are the YJL213w and YHL021c encoded proteins. Both YJL213w and YHL021c are hypothetical ORFs which are localized in the mitochondria.
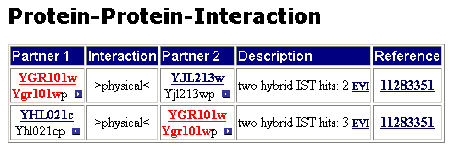
Figure 3: Interactions listed by MIPS.
Protein Function Analysis:
►WIT: Yields no information on PCP1.
►MIPS: Provides the Gene Ontology information obtained earlier.
CONCLUSIONS
The information in the searched databases serves to verify that PCP1 is involved in mitochondrial organization and biogenesis and that it is localized in the mitochondrion. We also obtained some additional information about the protein such as the molecular mass and pI.
FURTHER ANALYSIS
Even though PCP1 is a well characterized gene, and we already know that it is involved in mitochondrial organization, we could still perform further experiments to explore the precise role of the gene in the life of Saccharomyces cerevisiae. Determining the 3D structure of the protein and the nature and location of the active sites would be of tremendous help in the investigation. We could make a head start in this direction by doing sequence analysis and conducting experiments to try and verify any structural predictions.
REVIEW
YGR102C is a hypothetical ORF which, like PCP1, is located on chromosome VII in Saccharomyces cerevisiae. Its cellular component, biological process, and molecular function are unknown. Earlier, I had predicted that YGR102C is probably a nucleolar protein involved in rRNA processing.
PROTEOMIC ANALYSIS:
Structure:
►Protein Data Bank (PDB): A search for the structure of the YGR102C encoded protein yields no results.
►PROWL: Sequence analysis calculates the molecular mass of YGR102Cp as 20773.960 Da, and the isoelectric point pI as 9.7.
Protein Interaction Networks:
►Yeast Two-hybrid: According to this database YGR102C has no interactions.

Figure 4: YGR102C has no interactions.
► The Database of Interacting Proteins (DIP): The graph below depicts the protein-protein interactions for YGR102C.

Figure 5: Graph obtained from the DIP which shows the protein-protein interactions for YGR102C.
In this figure, the red node denotes the YGR102C protein. It directly interacts with only one other protein which is shown as the central node. This is the FUN12 protein. According to the Saccharomyces Genome Database (SGD), FUN12 is involved in translational initiation, and is localized in the cytosolic small ribosomal subunit (sensu Eukarya).
►The Benno Schwikowski figures: YGR102C is not one of the proteins in the interaction map.
►MIPS: No interactions were listed for the YGR102C encoded protein.
Protein Function Analysis:
►WIT: Yields no information on YGR102C.
CONCLUSIONS
As mentioned before, I had previously speculated that YGR102C could be a nucleolar protein involved in rRNA processing. However, the limited data that I've obtained from proteomic databases does not really support this claim. The only protein that appears to interact with the YGR102C encoded protein is the one encoded by FUN12. But FUN12 is not involved in translational initiation and neither is it localized in the nucleolus.
FURTHER ANALYSIS
Analysis of YGR102C is especially difficult at this stage because most of the genes that it interacts with or exhibits sequence similarity with are as yet unannotated. We should design experiments to test a hypothesis about the gene's molecular function and biological process. We could use some sort of fluorescent marker which could bind to the protein and help to find the cellular location of YGR102C. Determining the 3D structure of the protein and the nature and location of the active sites would be of tremendous help in the analysis.
REFERENCES:
Campbell, Malcolm A., Laurie Heyer. 2002. Discovering Genomics, Proteomics, and Bioinformatics. Benjamin Cummings, San Francisco, CA.
Database of Interacting Proteins. <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3> Accessed 20 November 2003.
Protein Data Bank. <http://www.rcsb.org/pdb/index.html> Accessed 20 November 2003.
PROWL. <http://129.85.19.192/prowl/proteininfo.html> Accessed 20 November 2003.
MIPS Comprehensive Yeast Genome Database <http://mips.gsf.de/genre/proj/yeast/index.jsp> Accessed 20 November 2003.
Saccharomyces Genome Database. <http://www.yeastgenome.org/> Accessed 20 November 2003.
Schwikowski, Benno, Peter Uetz, and Stanley Fields. 2000. A network of protein-protein interactions in yeast. Nature Biotechnology. 18: 1257-1261.
What Is There Database. <http://wit.mcs.anl.gov/WIT2/CGI/index.cgi> Accessed 21 November 2003.
Y2H Results Full Database. <http://portal.curagen.com/extpc/com.curagen.portal.servlet.PortalYeastList?modeIn=List> Accessed 20 November 2003.
Send questions, comments and suggestions to: pakarnik@davidson.edu