This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Annotated and Non-annotated Genes: SKI6 and YGR176W.
"Yeast strains carrying a 1.5 X 10(6)-dalton double-stranded RNA in virus-like particles secrete a protein toxin which is lethal to strains not carrying this species of double-stranded RNA. We find that recessive mutations in any of four chromosomal genes result in the superkiller phenotype, i.e., increased secretion of killer toxin activity by strains carrying the killer genome" (Toh-E et al., 1978).
Introduction and Purpose
With this web page I hope to elucidate the the roll SKI6 plays in the life of yeast and, through the use of web tools, explore the possible roles of the ORF sequence, YGR176W. There are many sources of genomic and bioinformatic data available on the internet; through this project, I will gain and provide insight on how to find and utilize these resources.
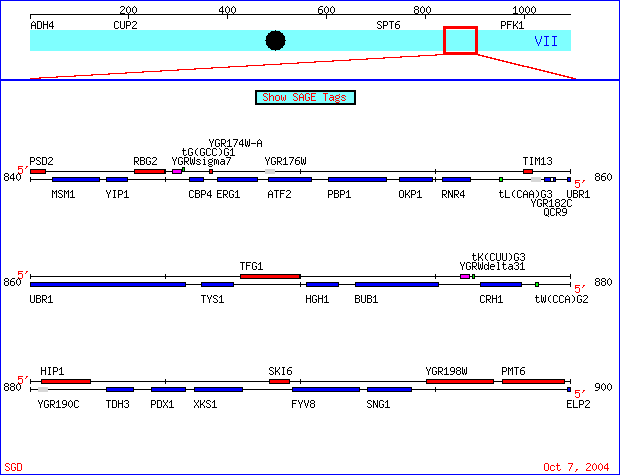 |
Figure 1. The genes I chose are located on the watson strand of chromosome VII of the S288C strain of Saccharomyces cerevisiae genome, coordinates 840000 - 900000. SKI6 (also known as ECM20, RRP41 and systematically YGR195) has been well characterized and documented. Conversely, specific information regarding the biological process, molecular function and
cellular component of YGR176W is unknown. SGD Chromosomal Features Map. New Gene View http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?chr=7&beg=880000&end=920000
|
Background Information
Saccharomyces cerevisiae is commonly known as baker's, brewer's, or budding yeast. It is a unicellular eukaryotic organism that serves as a model system for studying genomics; this is a result of the ease of genetic manipulations, high availability, rapid growth rate and versatile DNA transformation system (Sherman, 2002). |
 |
| |
Figure 2. Images of yeast (Saccharomyces cerevisiae) from University of Kent Biosciences. |
Annotated Gene - SKI6
The Saccharomyces Genome Database is the best place to begin if you want to discover information about the molecular biology and genetics of yeast. This scientific database is comprehensive and accessible. The authors conveniently list links to other resources, providing a wealth of information about any specific gene. The Basic Information Page for the verified ORF, SKI6, can be accessed at this address. <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=ski6>
The nucleotide sequence (741 bases) for SKI6 is available here in FASTA and GCG format or at NCBI with accession number NC_001139.
This gene is composed of 246 amino acids, this sequence can also be found in FASTA and GCG format or at NCBI P46948.
NCBI's Enrez Gene page for SKI6 provides a summary of the gene's function and helpful links.
Gene Ontology
This consortium wanted to universalize the terminology used to describe the functions of a protein. In order to do this, they decided to use three terms for why? what?and where? (Campbell, 2003).
The Gene Ontology web site describe the terms in the following manner:
"Biological Process - broad biological goals, such as mitosis or purine metabolism, that are accomplished by ordered assemblies of molecular functions.
Molecular Function - the tasks performed by individual gene products; examples are carbohydrate binding and ATPase activity.
Cellular Component - subcellular structures, locations, and macromolecular complexes; examples include nucleus, telomere, and origin recognition complex." |
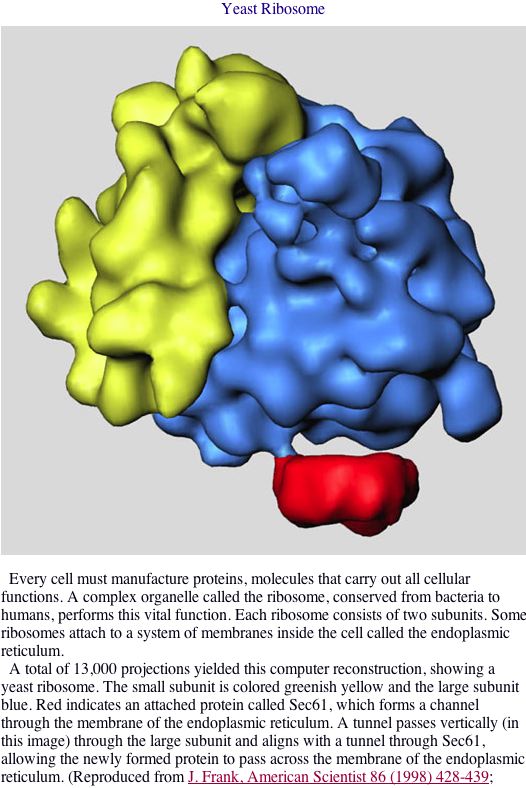 |
|
Figure 3. SKI6 is intricately involved in the ribosomal processes. ASCB. |
Biological Process
SKI6 plays a role in ribosome assembly and 3'-5' mRNA decay. A review paper explains SKI6 is, "homologous to the E. coli 3' 5' exonuclease RNase PH, a phosphorolytic nuclease that uses phosphate as an attacking group and releases nucleotide-5'-diphosphates. Recombinant Rrp41p [SKI6] and Mtr3p show a processive, phosphorolytic 3' 5' exonuclease activity in vitro" (Venema and Tollervey, 1999). Also, SKI6 plays a role in mRNA catabolism, defined as, "the breakdown of messenger RNA, which is responsible for carrying the coded genetic 'message', transcribed from DNA, to sites of protein assembly at the ribosomes" (GO).
Molecular Function
SKI6 is important on the molecular level because the function is specifically, 3'-5'-exoribonuclease activity, which GO defines as the "catalysis of the sequential cleavage of mononucleotides from a free 3' terminus of an RNA molecule." The process of RNA degradation is much more efficient when the strand can be dissolved from both directions simultaneously.
Cellular Component
SKI6 is a component of the exosome that makes the 3'-5' degradation of mRNA more efficient.
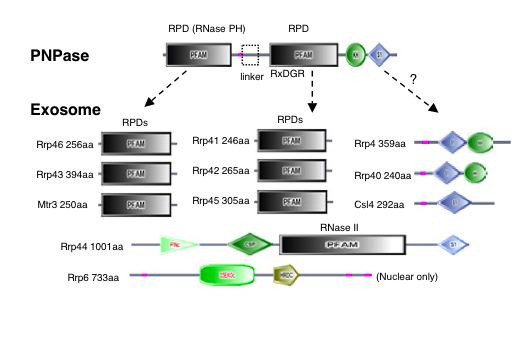 |
| Figure 4. "Domain architectures for the exosome core and PNPase. Domains are taken from SMART (colored shapes) or Pfam (boxes). Regions of low sequence complexity are shown in pink" (Aloy et al., 2002) |
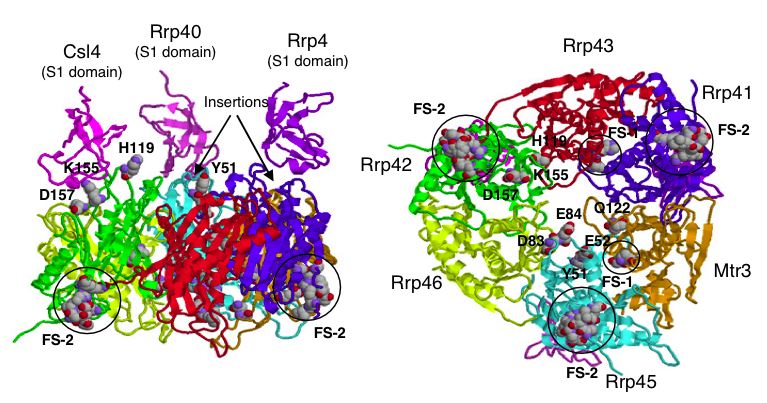 |
| Figure 5. A proposed 3-D model of the yeast exosome core. Notice the position of SKI6 (Rrp41) and the indication that it is a functional site for binding. For details on model creation see Fig5 in (Aloy et al., 2002). |
Mutant Alleles and Phenotypes
The amount of information available about mutant forms of SKI6 is quite limited, but enough resources can be found to conclude that SKI6 plays a unique and important role in the life of yeast. SGD had a small section of free text saying, "deletion mutation are inviable and mutants show superkiller phenotype, improved translation of non-poly(A) mRNA, abnormal 60S ribosomal subunits and defective 3' processing of 5.8S rRNA; a Tn3 insertion into this gene causes hypersensitivity to the cell surface polymer perturbing agent calcofluor white." The genetic footprinting of SKI6 deletions shows severe growth defects on YPD, NaCl, lactate, and minimal media.
Conservation
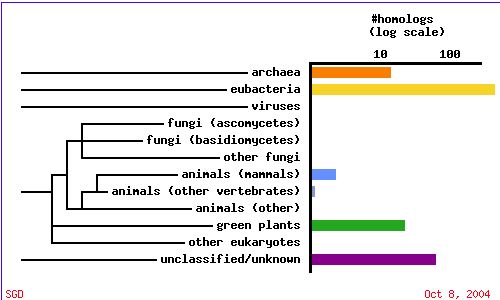
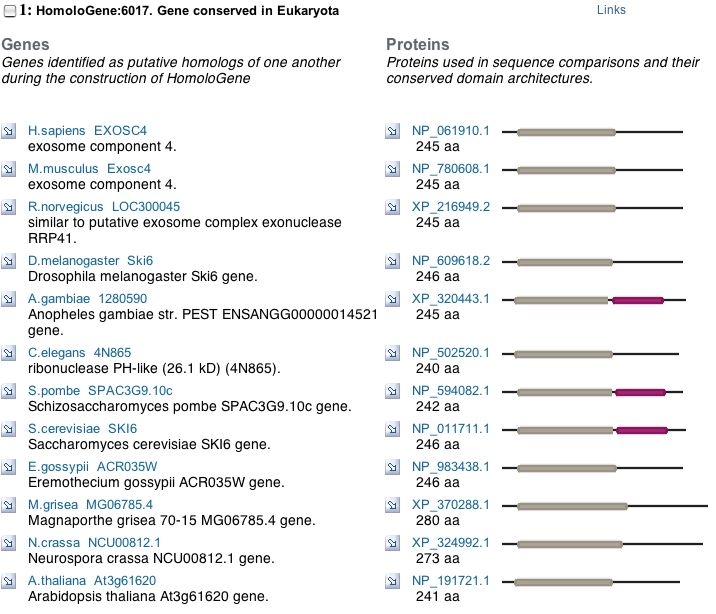
A PSI-BLAST shows that SKI6 is highly conserved and HomoloGene displays the specific conserved domains for a variety of organisms, indicating an important evolutionary function.
Non-annotated Gene - YGR176W
Though SGD has limited information about dubious ORFs, the database is still a good starting point for analysis. <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YGR176W>
Swiss-Prot also has a page for YGR176W at <http://seq.yeastgenome.org/cgi-bin/nph-blast-fungal.pl?name=YGR176W&reason=2>
The nucleotide sequence (348 bases) for YGR176W is available here in FASTA and GCG format.
This gene is composed of 116 amino acids, this sequence can also be found in FASTA and GCG format.
With a greater background in chemistry, BIND interaction information could be helpful. <http://bind.ca/Action?textquery=RecordType:%20(interaction%20complex%20pathway%20)+AND+YGR176W>
I began with the ORF sequence and used BLAST, Kyte-Doolittle Hydopathy, and conservation/homolog sources to explore the role of this gene. I first found that a system deletion is viable, which indicates that the function of YGR176W may be nonessential or redundant.
 |
 |
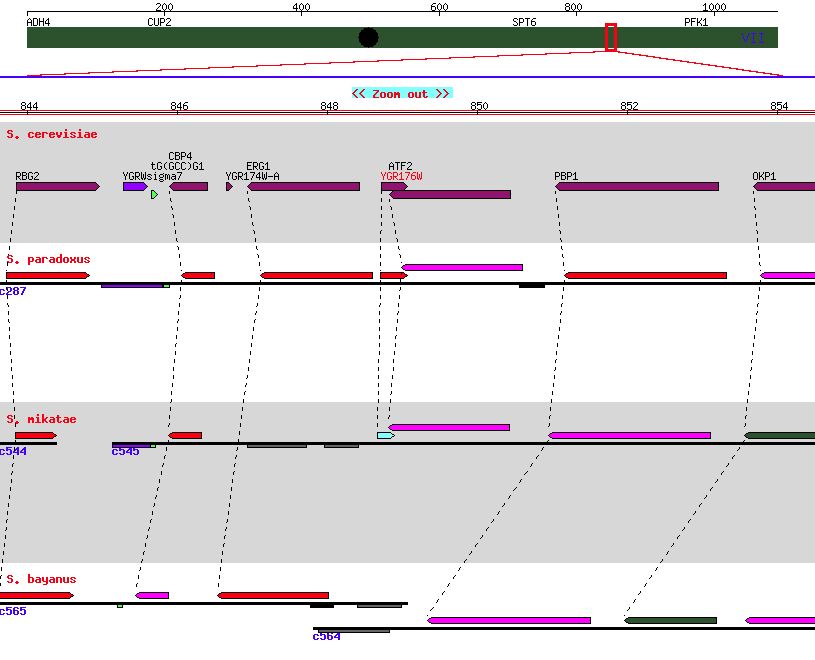
| Figure 6. A genome comparison for other closely related yeasts indicating minor conservation. SGD. |
|
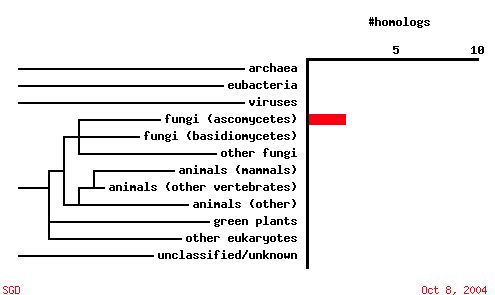 |
| Figure 7. A PSI-BLAST analysis shows that in contrast to SKI6, YGR176W ORF sequence is not conserved among other organisms. |
Since fungi appeared to be the only organism with conservation, I preformed a BLAST search to compare the sequences.
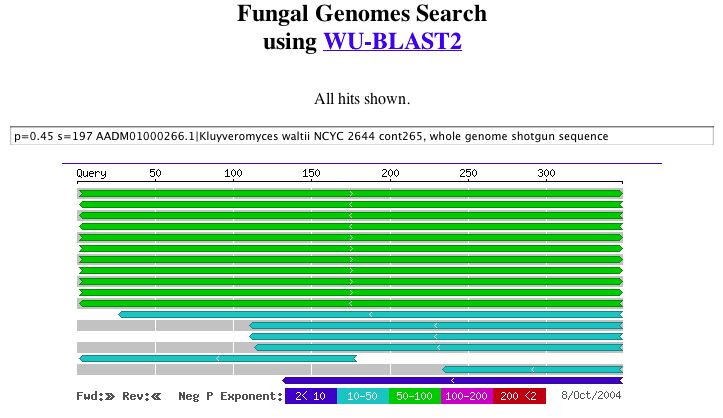 |
 |
| Figure 8. A potential role related to alcohol transferase can be seen in these results. WU-BLAST2. |
|
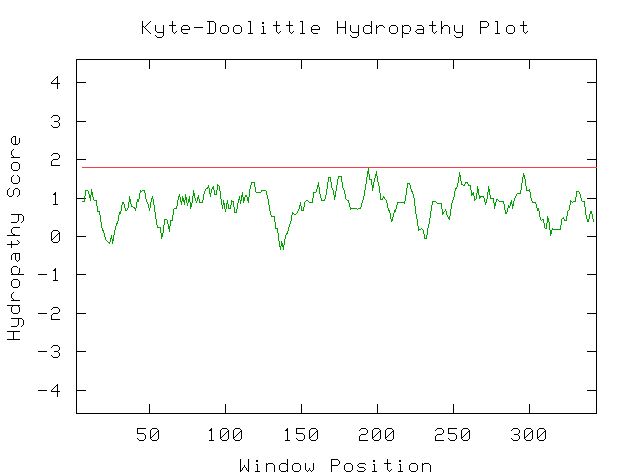 |
| Figure 9. This Kyte-Doolittle plot shows that YGR176W a positive hydropathy for most of its length. However, none of the peaks are wide or have a hydropathy greater than 1.8, indicating that it is not likely to be a transmembrane protien. There is not enough information to determine if this protein would be found in an aqueous environment. |
 |
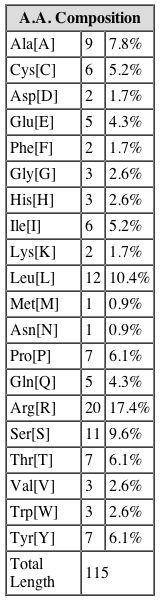 |
Figure 10. Similarly expressed genes in response to alpha-factor. This analysis illustrates the roles of genes that are similar to YGR176W and provides some basis to form conclusions about the function of the dubious ORF. From SGD Alpha-factor Analysis. |
Amino Acid Composition of YGR176W. SGD. |
I utilized the GO term mapper with the similar genes found above <http://db.yeastgenome.org/cgi-bin/GO/goTermMapper>.
Prediction: I believe that my non-annotated gene is present in the cytoplasm and plays a roll in transfer of materials within and between cells.
Links and References
Allmang C, et al. (1999) The yeast exosome and human PM-Scl are related complexes of 3' --> 5' exonucleases. Genes Dev 13(16):2148-58 <http://www.genesdev.org/cgi/content/full/13/16/2148>
Aloy, P., Cicarelli, F.D., Leutwein, C., Gavin, A., et al. (2002) EMBO Rep. 628-635. A complex prediction: three-dimensional model of the Yeast exosome. <http://www.nature.com/cgi-taf/DynaPage.taf?file=/embor/journal/v3/n7/full/embor123.html>
Cherry JM. et. al. (1997) Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67-73.
Giaever G, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418(6896):387-91
Mitchell, P. , Petfalski, E. , Shevchenko, A. , Mann, M. and Tollervey, D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3'5' exoribonuclease activities. Cell, 91, 457-4
Ozaki-Kuroda K, Yamamoto Y, Nohara H, Kinoshita M, Fujiwara T, Irie K, Takai Y (2001) Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol Cell Biol 21(3):827-39 <http://mcb.asm.org/cgi/content/full/21/3/827?view=full&pmid=11154270>
Sherman 2002 Getting Started with Yeast. Methods Enzymol. 2002;350:3-41
Toh-E, A., Guerry, P., and Wickner. (1978) Chromosomal superkiller mutants of Saccharomyces cerevisiae. J Bacteriol. 136 (3): 1002–1007 http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=218536&action=stream&blobtype=pdf
Dolinski, K. et. al. 2003. Saccharomyces Genome Database. <http://www.yeastgenome.org> Accessed 4 Oct. 2004.
Gene Ontology Software Group. 2003. Gene Ontology Database.<http://www.godatabase.org/dev/database> Accessed 6 Oct. 2004.
NCBI. 2003. National Center for Biotechnology Information. <http://www.ncbi.nih.gov> Accessed 5 Oct.2004.
SwissProt. 2003. Swiss-prot protein knowledgebase. <http://us.expasy.org/sprot> Accessed 6 Oct. 2004.
Davidson College Home
Biology at Davidson
Genomics at Davidson
Sheena’s Genomics Home
Questions or Comments? Email: shbossie@davidson.edu
© Copyright 2004 Department of Biology, Davidson College, Davidson, NC 28036














