Introduction
In My Favorite Yeast Gene, I described the annotated PDI1 gene and explored available information on the neighboring YCL047C hypothetical ORF, which is entirely uncharacterized. In my follow-up My Favorite Yeast Expression, I examined the expression patterns of PDI1 and YCL047C, in order to further explore the role of PDI1 and to refine my proposed hypothetical function for YCL047C. This page will utilize online proteomics databases, in order to investigate the roles which the protein products of PDI1 and YCL047C play within Saccharomyces cerevisiae cells.
Review of PDI1
∙ PDI1 is located within a small region of the Crick strand of Saccharomyces cerevisiae chromosome 3 (coordinates 50221-48653).
∙ PDI1 encodes an essential protein disulfide isomerase. This protein product catalyzes the formation of native disulfide bonds in secretory proteins, an important factor leading to the overall structure of these proteins.
∙ The folding of secretory proteins occurs within the lumen of the endoplasmic reticulum. Consequently, the protein product of PDI1 is also found in this region.
∙ The PDI1 protein product contains two thioredoxin-like domains, each with a copy of the active site sequence motif CGHC. The two cysteines within these active sites possess the ability to cycle between a reduced and an oxidized state. In the oxidized state, the two cysteines form an intramolecular disulfide bond that enables PDI1 to convert a pair of sulfhydryl groups in a polypeptide substrate into a disulfide bond.
PDI1 Protein Data Analysis
Protein Structure
A search of PDB yielded no results for “PDI1.” Similarly, no results were found for any of the genetic homologs (within S. cerevisiae) of PDI1; however, a query for “PDI” resulted in fourteen structures. Of these, three hits were for protein disulfide isomerase molecules in humans. The PDB image below is of the 1MEK human protein disulfide isomerase molecule.
Figure 1. Human Protein Disulfide Isomerase: 1MEK . This molecule, found in humans, is somewhat homologous to PDI1 in S. cerevisiae. 1MEK has a strong amino acid sequence similarity with PDI1 in the thioredoxin-like domains; however, 1MEK is only 120 amino acids long, whereas PDI1 is 522 amino acids long. Image from: <http://www.rcsb.org/pdb/cgi/explore.cgi?pid=278191100830460&page=0&pdbId=1MEK>. Permission pending.
Can't see the image? Click here to download the FREE Chime Plugin.
This molecule, while it shares a great deal of sequence similarity in the thioredoxin-like domains, is not a very accurate indicator of the shape of the rest of the PDI1 molecule, as 1MEK is only 120 amino acids long (while PDI1 is 522 amino acids long). However, this picture has been included because of the similarity in the thioredoxin-like active sites. |
A search for “PDI1” on the PROWL website yielded a sequence analysis result for molecular mass (54041.637 Da) and isoelectric point (7.7). These results will be useful in examining the 2D gel for S. cerevisiae.
Protein Function & Interactions
| According to the PROWL site, the molecular mass of the PDI1 protein product is 54041.637 Da and the isolectric point is 7.7; however, as indicated below, I was unable to find any information for PDI1 on the 2D gel. |
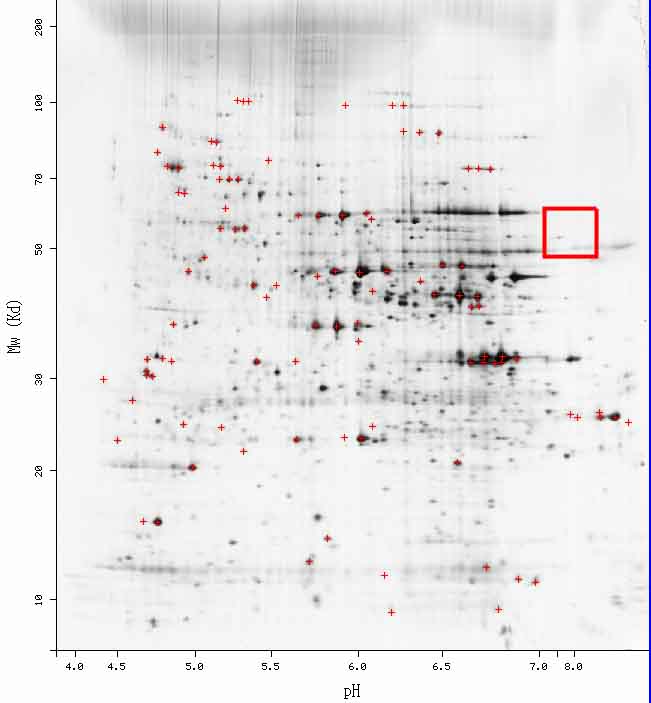
Figure 2. Saccharomyces cerevisiae 2D Gel. According to the PROWL website, PDI1 should be somewhere in the red box; while there are some areas of darkness, currently, no information has been plotted in this area for PDI1.
PDI1 is a very active protein. The following table lists eight proteins with which PDI1 is known to associate.
Figure 3. Proteins with which PCI1 is Known to Associate. This table illustrates the variety of proteins, representing a spectrum of processes, with which PDI1 associates. This variety leads one to assume that PDI1 may have broad-reaching effects, all of which are not yet fully understood. Image from: http://biodata.mshri.on.ca/. Permission pending.
| This table clearly shows that PDI1 interacts with many proteins, associated with many processes. This result leads one to assume that PDI1 may have broad-reaching effects - this result is consitent with the known function of PDI1 (an essential protein disulfide isomerase which catalyzes the formation of native disulfide bonds in many proteins). |
A search of the DIP database for PDI1 yielded the following image.
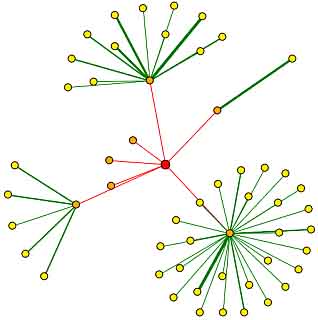
Figure 4. Protein Interaction Map for PDI1. The red node in the center of this image represents the root node – PDI1. The orange nodes represent the first shell of nodes (the proteins which interact directly with PDI1) and the yellow nodes represent the second shell of nodes (proteins two edges away from the root node – PDI1). Edges drawn in green represent core interactions that have been verified by one or more computational verification methods, while red edges represent unverified results attained via high-throughput analysis. Image from: <http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=4978>. Permission pending.
The orange nodes in this figure each represent one of the seven proteins with which PDI1 has been proven to interact directly. These first shell nodes include: EST1, CDC11, TAL1, MDH2, YER189w, SGN1, and CKA1. The functions of these proteins are briefly described below. |
EST1 – RNA-associated factor involved in telomere length regulation as the recruitment subunit of the telomerase holoenzyme; has a possible role in activating Est2p-TLC1-RNA bound to the telomere (SGD)
CDC11 – Component of the septin ring of the mother-bud neck that is required for cytokinesis (SGD)
TAL1 – Transaldolase – enzyme in the pentose phosphate pathway (SGD)
MDH2 – Cytoplasmic malate dehydrogenase – catalyzes interconversion of malate and oxaloacetate; involved in gluconeogenesis and the glyoxylate cycle (SGD)
YER189w – Uncharacterized ORF (SGD)
SGN1 – Cytoplasmic RNA-binding protein which contains an RNA recognition motif (RRM); may have a role in mRNA translation (SGD)
KA1 – Alpha subunit of protein kinase CK2 (SGD)
Again, the fact that these proteins have a variety of functions may, at first, be somewhat perplexing; however, this data is consistent with the known function of PDI1. As a protein disulfide isomerase, this protein is involved in the formation of native disulfide bonds. Thus, PDI1 may act on a variety of proteins. |
The interaction diagram produced by Schwikowski, et al., did not include PDI1.
No information about protein interactions of PDI1 was given in the results of the yeast two-hybrid analysis, nor was PDI1 included in the addendum, published after the original study (Uetz, et al. 2000).
The MIPS Protein-Protein Interaction database lists four proteins that interact with PDI1 (Figure 5).
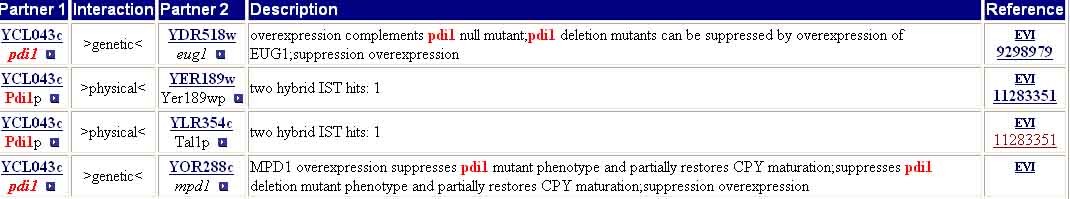
Figure 5. MIPS Protein-Protein Interactions of PDI1. A query of the MIPS Protein-Protein Interactions database for “PDI1” resulted in hits for EUG1, YER189W, TAL1P, and MPD1. Image from: <http://mips.gsf.de/proj/yeast/CYGD/interaction>. Permission pending.
Of the four results returned by the MIPS Protein-Protein Interaction database, two hits (EUG1 and MPD1) are for homologous proteins with thioredoxin-like domains and two hits (YER189W and TAL1P) are for proteins which were described above (as results in the DIP database search). These results support the interaction of PDI1 with YER189W and TAL1P proteins, found in earlier analyses. In addition, the interaction with homologous EUG1 and MPD1 proteins makes sense, as all of these proteins perform similar functions, and their interactions have to do with control mechanisms (feedback loops affecting protein production). |
The two cysteines within these active sites possess the ability to cycle between a reduced and an oxidized state. In the oxidized state, the two cysteines form an intramolecular disulfide bond that “in principle, enables PDI to convert a pair of sulfhydryl groups in a polypeptide substrate into a disulfide bond” (Figure 6).
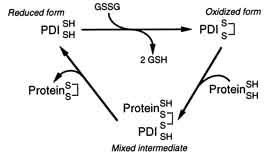
Figure 6. Reaction Cycle for the Oxidation of a Nascent Polypeptide Catalyzed by PDI. This figure illustrates an oxidation reaction in which the intramolecular disulfide bond of the CGHC motif is transferred to a pair of sulfhydryls in a substrate polypeptide. Image from: (Holst 1997). Permission pending.
Enzymes and Metabolic Pathways provided no new information.
A search of the KEGG database produced no new information.
A search of the ExPASy database provided no new information.
Review of YCL047C
∙ YCL047C is located within a small region of the Crick strand of Saccharomyces cerevisiae chromosome 3 (coordinates 44437- 43661).
∙ A BLASTp search resulted in a strong hit for the AFR721Wp protein in Eremothecium gossypii. Within this protein, there is a cytidylyltransferase conserved domain. Cytidylyltransferases form the critical intermediates in the biosynthesis of lipids and complex carbohydrates.
∙ A conserved domain search of the YCL047C sequence resulted in only one hit – the nicotinic acid mononucleotide adenylyltransferase domain, used in coenzyme metabolism.
∙ The Kyte-Doolittle hydropathy plot predicts the presence of two transmembrane domains – one near amino acid 65 and another near amino acid 108.
∙ On my previous page, I hypothesized that YCL047C is a transmembrane functional protein involved in biosynthesis (metabolism) of anabolic carbohydrates.
YCL047C Protein Data Analysis
Protein Structure
A search of PDB for “YCL047C” yielded no results. Similarly, neither the homologous “AFR721W” protein (found in Eremothecium gossypii), nor the homologous “SPAC694.03” protein (found in Schizosaccharomyces pombe) produced any results.
A search for “YCL047C” on the PROWL website yielded a sequence analysis result for molecular mass (29654.301 Da) and isoelectric point (8.5). These results will be useful in examining the 2D gel for S. cerevisiae.
Protein Function & Interaction
| According to the PROWL site, the molecular mass of the YCL047C protein product is 29654.301 Da and the isolectric point is 8.5; however, as indicated below, I was unable to find any information for YCL047C on the2D gel. |
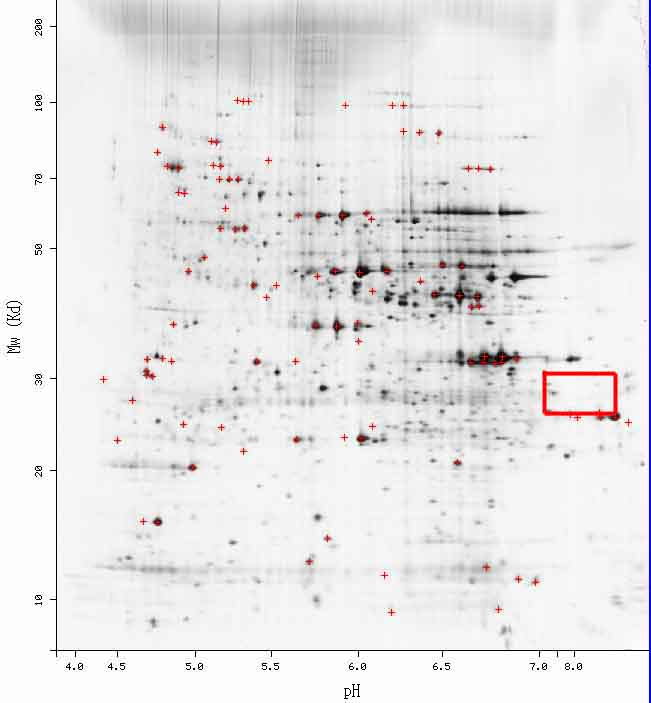
Figure 7. Saccharomyces cerevisiae 2D Gel. According to the PROWL website, YCL047C should be somewhere in the red box; while there are some areas of darkness, currently, no information has been plotted in this area for YCL047C.
The image below shows the insertion of an mTn element into YCL047C ORF. This insertion caused a gene “knockout."
![]()
Figure 8. YCL047C Knockout Yeast. This image shows hit in the TRIPLES database for the mTn insertion in the YCL047C ORF. Image from: <http://ygac.med.yale.edu/triples/>. Permission pending.
| The results presented for this hit provide little substantive information. Two possible insertion points are given – one "in frame" and one in the "3’ region"; however, no information on phenotypes, localization, or expression is provided. |
A search of the DIP database for “YCL047C” did yield a hit for my hypothetical ORF; however, no new information was presented in any of the links and a protein interaction map was not available.
The interaction diagram produced by Schwikowski, et al., also failed to include YCL047C.
No information about protein interactions of YCL047C was given in the results of the yeast two-hybrid analysis, nor was YCL047C included in the addendum, published since the original study (Uetz, et al. 2000).
Enzymes and Metabolic Pathways provided no new information.
A search of the KEGG database produced no new information.
A search of the ExPASy database provided no new information.
| Since there was virtually no new information produced in my searches of online proteomics tools, I will stand by my previous hypothesis. I predict that YCL047C is a functional protein involved in biosynthesis (metabolism) of anabolic carbohydrates. Since no conclusive evidence has been presented contrary to the Kyte-Doolittle hydropathy plot, I will stick with my original hypothesis that YCL047C codes for a transmembrane protein. |
Although I was able to find a limited amount of data concerning the YCL047C ORF in previous pages, the results of my searches of online proteomic databases were less than fruitful. In many cases, it was not that YCL047C did not produce a significant result, but that the YCL047C ORF was simply not included in the findings. Therefore, experimentation to begin to uncover the protein function of YCL047C need not consist of new and novel approaches to proteomic research; rather, current techniques can be used as a starting point. By performing the following basic experiments, we would have a solid basis of knowledge concerning the function of YCL047C. From here, new and novel approaches could be employed to further refine the function of the YCL047C protein product. First, the cellular component of YCL047C must be defined. The Kyte-Doolittle hydropathy plot for YCL047C suggests that the protein product may be a transmembrane protein. In order to test this hypothesis, a green fluorescent protein (GFP) antibody could be bound to the YCL047C protein product, thus, “tagging” the protein product. By monitoring this green fluorescent labeling, one should be able to visualize the location of the YCL047C protein product throughout an organism’s life cycle. Next, I would attempt a yeast two-hybrid analysis, in order to discern some protein-protein interactions. Using YCL047C as the bait protein (the protein bound to the DNA Binding Domain), I predict that the reporter (His3) gene would be transcribed most often when prey proteins consisted of proteins involved in the biosynthesis (metabolism) of anabolic carbohydrates bound to the Activation Domain. This result would imply that YCL047C interacts with proteins involved in the biosynthesis of anabolic carbohydrates pathway, the basis of my current functional hypothesis. Finally, I would create a knockout strain of YCL047C and study the effects that this “knockout” would have on the organism, in various environmental conditions. The results of this knockout could produce organisms which are inviable in different conditions. If this is the case, then the YCL047C protein must be vital to the organism under these environmental conditions. Other pheonotypic variants may also be noted under varying conditions. By analyzing the changes from the wildtype, presented by the knockout, we may be able to learn more about the function of YCL047C. |
References
[DIP] Database of Interacting Proteins. 2003. <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>. Accessed 2004 Nov 19.
Dolinski K, et al. 2004. Saccharomyces Genome Database. <http://www.yeastgenome.org>. Accessed 2004 Nov 19.
MIPS Comprehensive Yeast Genome Database. 2003. <http://mips.gsf.de/genre/proj/yeast/index.jsp>. Accessed 2004 Nov. 19.
Ross-Macdonald P, et al. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402: 413-8.
Schwikowski B, et al. 2000. A Network of Protein-Protein Interactions in Yeast. Nature Biotechnology 18: 1257-1261.
Uetz P, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623-7.
