
*This page was created as an undergraduate assignment for a Genomics course at Davidson College*
YNK1 and YKL069W
My Favorite Yeast Proteins
In my previous two web pages, I have analyzed my favorite yeast gene YNK1 and its neighboring non-annotated ORF YKL069W based on there sequence and expression patters. I have concluded based analyzing the sequence and expression patterns for these genes that YNK1 appears to be correctly as a Nucleoside diphosphate kinase. While I currently predict that YKL069W is a non-membrane protein found near ribosomes that is involved in protein biosynthesis, and it is highly repressed during histone depletion and induced during nitrogen depletion, suggesting it may be involved in nitrogen freeing or storing processes.
I will know analyze these YNK1 and YKL069W by exploring and studying their protein structures found in various proteomics databases.
Basic Protein Information for YNK1


Figure 1: Protein sequence information for YNK1. This information comes directly from the Saccharomyces Genome Database. Permission for image pending.
Protein Data Analysis for YNK1 (nucleoside diphosphate kinase)
I first searched Protein Data Bank for the structure of YNK1. I could not find the structure of any nucleoside diphospahte kinase homologs from Saccharomyces cerevisiae. From here I began to look for other nucleoside diphospahte kinases that had sequence conservations. No well conserved sequences were found to have PDB files.
YNK1 Function and Interaction
Searching for more information on YNK1 brought me to the DIP database. The DIP database has information on protein interactions.

Figure 2: Protein interactions map of YNK1. The red node in the middle represents YNK1, while the orange nodes (first shell) represent the proteins that are thought to interact directly with YNK1. The yellow node proteins (second shell) interact directly with the orange node proteins. The green edges show protein interactions that have been confirmed by one or more method, while red edges have only been predicted based on on computational model.
The 8 proteins that are located in the first shell are: Hhf1p, Sen15p, tem1p, Rpa135p, Kap95p, Srp1p, Yju2p, and Cmd1p.
These proteins have a wide range of functions as listed below:
Hhf1p- histone protein; core histone for chromatin assembly
Sen15p- subunit of tRNA splicing endonuclease
Tem1p- GTP binding protein; termination of the m-phase
Rpa135p- RNA polymerase I subunit A135
Kap95p- Karyopherin beta; forms dimer with Srp1p. Mediates nuclear import of cargo proteins
Srp1p- Karyopherin alpha; forms dimer with Kap95. Believed to be important in protein degredation.
Yju2p- involved in mRNA splicing
Cmd1p- Calmodulin; Ca++ binding protein
All 8 of these proteins directly interact with YNK1. If you look at these proteins, the interactions make sense. Each of these proteins is found in the same cellular location (nucleus and cytoplasm), and most are involved in nucleic acid biosynthesis, except Kap95p, Srp1p, and Cmd1p. However due to the nature of YNK1 it has been thought to have the ability to phosphorylate other proteins and may interact with these proteins in this manner. |
To determine if any other interaction with this protein existed, a search of the Yeast Grid website was conducted. This table shows the proteins that YNK1 is thought to interact with. Upon looking closely you can determine that the proteins listed are the same proteins found in the first shell of nodes in the Protein interaction map found at the DIP database.
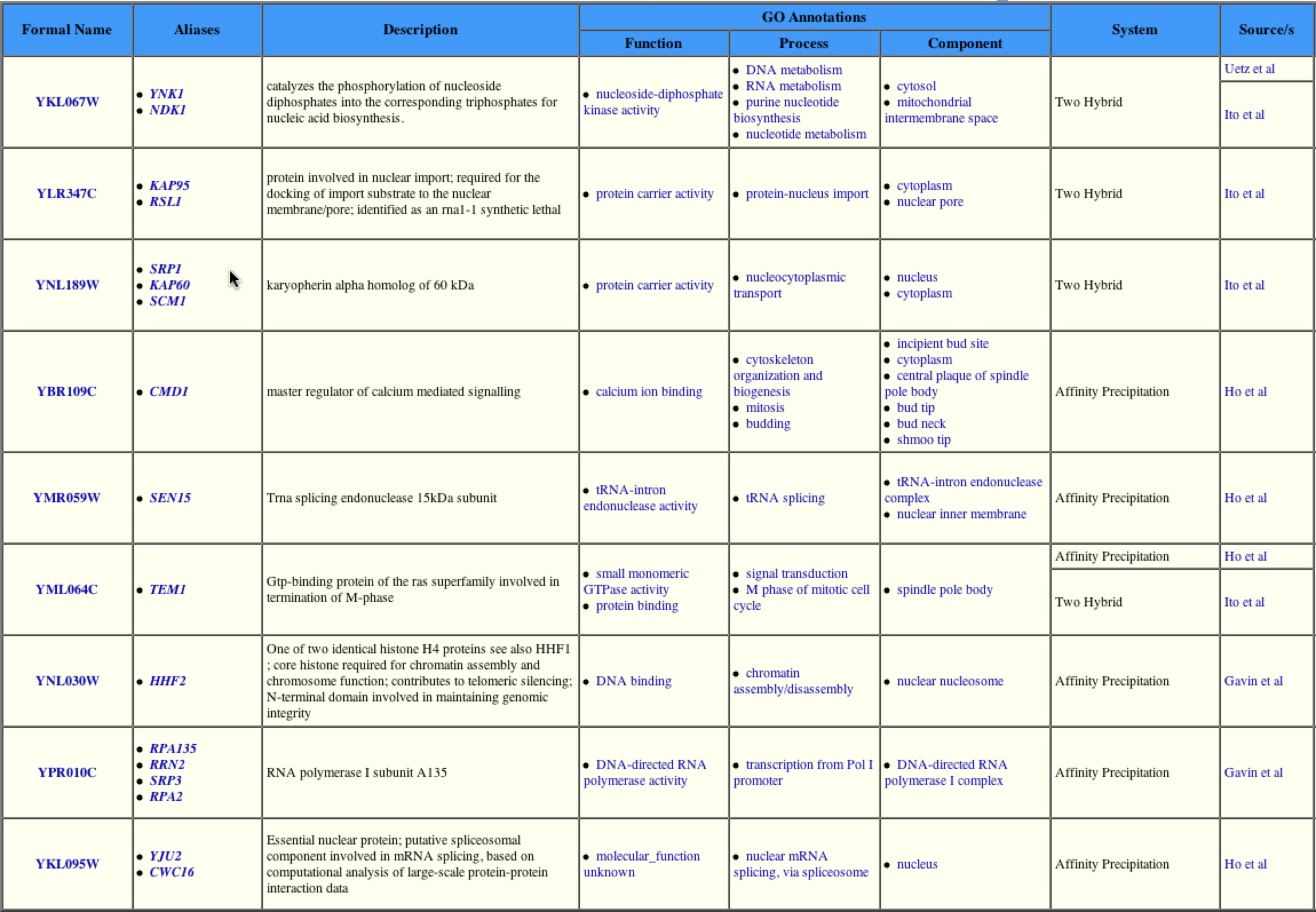
Figure 3: List of Proteins know to associate with YNK1. This table shows that YNK1 interacts with 8 different proteins excluding itself. Theser happen to be the same 8 found on the DIP interaction database. Image from: http://biodata.mshri.on.ca/. Permission pending.
Once again this shows that YNK1 interacts with several proteins directly involved in nucleic acid biosynthesis as well as a few proteins involved in other pathways. This is due YNK1 ability to phosphorylate other proteins, potentially turning on or off activity of these proteins. |
A search of the PROWL database showed that YNK1 had an isoelectric point of 8.6 and a molecular mass of 17166.71 Da. A search of the 2D gel revealed that YNK1 was not one of the proteins marked on the gel. With the information from PROWL it is possible to look at the 2D protein gel of S. cerevisiae and predict where YNK1 would be located. |

Figure 4: A 2D Gel Page of S. cerevisiae. The red box represents the hypothetical location of the YNK1 protein based on its pI (8.6) and its molecular mass (17166.7). There appears to be one dot inside the area of the red box, but it has not been identified.
Interaction information for YNK1 was also found at the MIPS protein-protein interaction database.
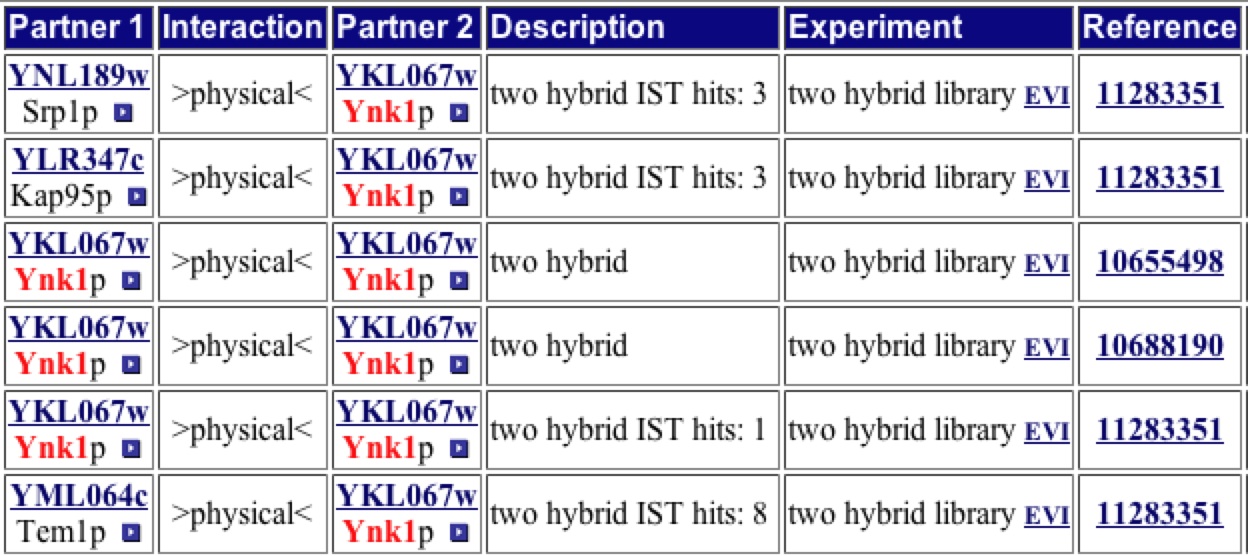
Figure 5 : MIPS protein-protein interaction of YNK1. Image from: <http://mips.gsf.de/proj/yeast/CYGD/interaction>. Permission pending.
The information found at the MIPS protein-protein interaction database only provides more evidence that YNK1 interacts with Srp1p, Kap95p, Tem1p, and itself.
|
YNK1 was included in the results of the yeast two-hybrid analysis, but the only interaction listed is between YNK1 and Ynk1 (Uetz, et al. 2000).
A search of the interaction diagram produced by Schwikowski, et al., does not include YNK1.
A search of the KEGG database showed that YNK1 or 2.7.4.6 (number code in these pathways) is involved in both the purine and pyrimidine pathways. This makes sense as YNK1 is involved in nucleic acid biosynthesis.
A search of the TRIPLES database provided no new information on YNK1.
The results obtained from searching several proteomics database only continue to affirm that YNK1 encodes a nucleside diphosphate kinase, which is involved in protein biosynthesis. The next step in gaining more understanding of this protein would be to crystalize the protein and determine its structure via X-Ray crystallography to determine the exact structure. This will also allow for a Jmol image to be made, making it easier to view and understand how YNK1 interacts with other proteins. |
Review of YKL069W
I currently predict that YKL069W is a non-membrane protein found near ribosomes that is involved in protein biosynthesis, and it is highly repressed during histone depletion and induced during nitrogen depletion, suggesting it may be involved in nitrogen freeing or storing processes. Also important to note that YKL069W also includes a GAF binding domain. GAF domains are 110 amino acids sequences that are often found in cyclic nucleotide phosphodiesterases (Zoraghia et. al 2004, Martinez et. al 2002). These domains often lead to the binding of a cyclic nucleotide. These GAF domains are found in 1400 proteins predicted by a non-redundant database (http://smart.embl-heidelberg.de), leading one to believe that the GAF domain may have other functions that have not be previously charactertised (Zoraghia et. al 2004).
Basic Protein information for YKL069W
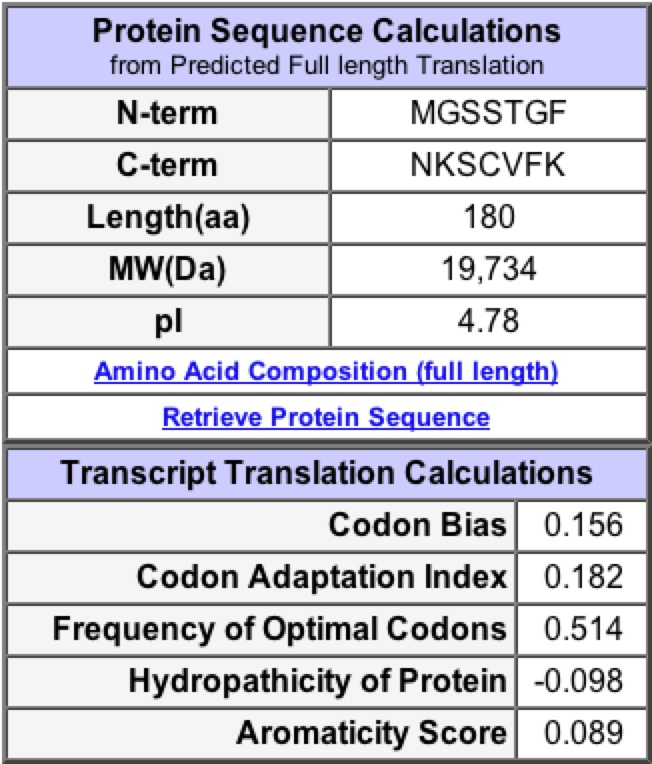

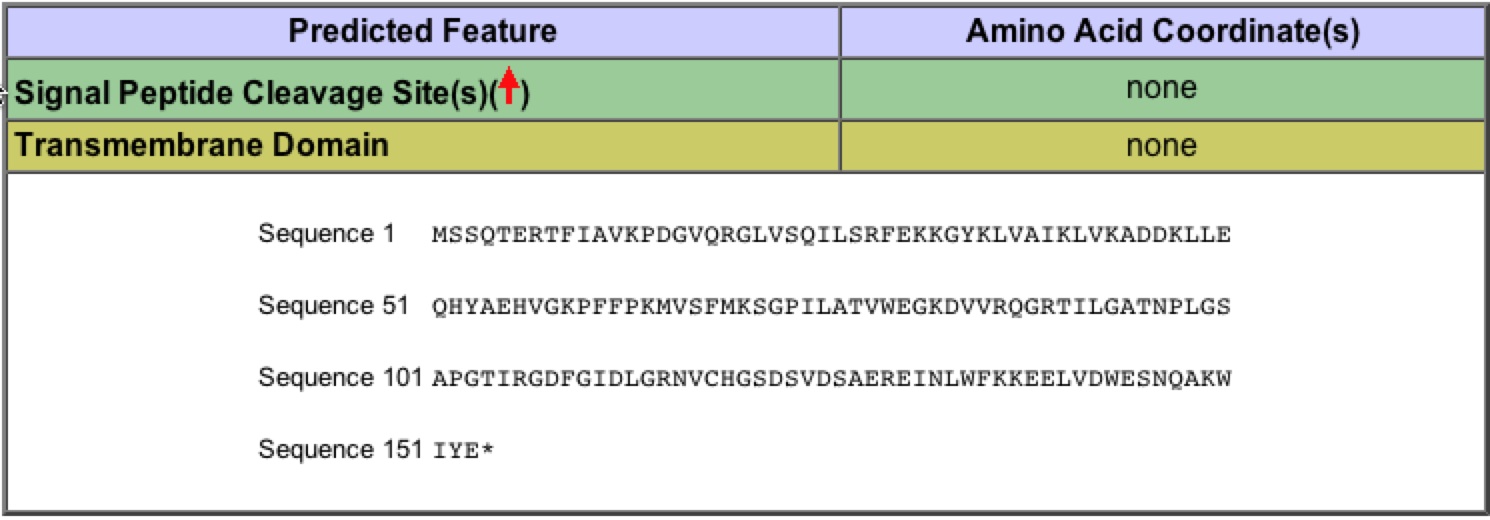
Figure 6: Protein sequence information for YKL069W. This information comes directly from the Saccharomyces Genome Database. Permission for image pending.
| The basic protein information from SGD, give general information about protein length (180 aa) or information about transmembrane domains. It also provides some information about hydrophobicity and aromaticity. |
Structure of YKL069W
There were hits for YKL069W in the Protein Data Bank, and no homologous proteins could be found. YKL069W does contain a GAF domain. I have found a synthetic structure of a GAF domain. The GAF domain of the YKL069W protein is highly conserved in sequence to this artificial GAF domain (1F5M), so they may look similar in shape.
| The synthetic GAF domain picture in the QuickPDB from Protein Data Bank, is 180 amino acids in length and by blast analysis (fig.7.) are the same 180 amino acids that comprise YKL069W. Another important thing to point out is that in the QuickPDB 1F5M is shown in a homodimer. Therefore I conclude that the QuickPDB of 1F5M is a very accurate representation of the structure of YKL069W when involved in a homodimer. |

Figure 7: Blast2 results from blasting YKL069W protein sequence against 1F5M's protein sequence found at PDB.
YKL069W Functions and Interactions
A search for YKL069W in the PROWL database provided information about molecular mass (19734.35 Da) and Isoelectric point (4.9). From these values one can determine the hypothetical location of YKL069W on the 2D page gel. This cannot be confirmed by at the 2D gel website, because a search for YKL069W provided no results. |

Figure 8: A 2D Gel Page of S. cerevisiae. The red box represents the hypothetical location of the YKL069W protein based on its pI (4.9) and its molecular mass (19734.35). There appears to be two dots inside the area of the red box, but these dots have not been identified.
A search of the DIP protein interaction map for YKL069W resulted in the following image:
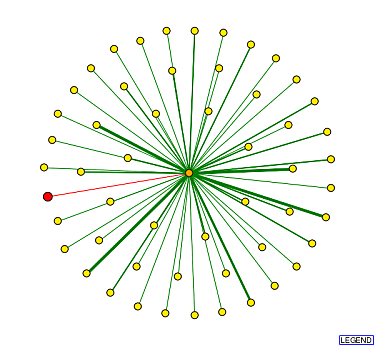
Figure 9: Protein interaction map of YKL069W. The red node in the middle represents YKL069W, while the orange nodes (first shell) represent the proteins that are thought to interact directly with YKL069W. The yellow node proteins (second shell) interact directly with the orange node proteins. The green edges show protein interactions that have been confirmed by one or more method, while red edges have only been predicted based on on computational model.
As one can see in this image, YKL069W is only hypothesized to interact with one protein. This one orange node represents Srp1p. One of the 8 proteins that directly interacted with YNK1. Srp1p is the karyopherin alpha unit in a dimer. It is believed to be involved in regulating protein degredation. |
A search of the MIPS protein-protein interaction database provided only one hit. It once again showed that YKL069W interacts with Srp1p (Karyopherin alpha) a protein that forms a dimer with Kap95. It is believed to be important in protein degredation. . |

Figure 10: Results from a YKL069W query at the MIPS protein protein interaction database. Image from: <http://mips.gsf.de/proj/yeast/CYGD/interaction>. Permission pending.
YKL069W was not included in the results of the yeast two-hybrid analysis, or the addendum 1 (Uetz, et al. 2000).
A search of the interaction diagram produced by Schwikowski, et al., does not include YKL069W.
A search of the KEGG database showed that YKL069W is not currently included in any pathways.
A search of the TRIPLES database provided no new information on YKL069W.
| Based on the limited knowledge gained from the search of the proteomics databases, I will hypothesize the function of my non-annotated ORF and protein/s. for the last time. I have no reason to believe that YKL069W represents a transmembrane protein as no new information has been found to contradict the Kyte-Doolittle hydropathy plot which suggested YKL069W was not an integral membrane protein. I still conclude that my protein may form a homodimer and be involved in signalling pathways directly related to protein biosynthesis. It is highly repressed during histone depletion and induced during nitrogen depletion. This suggests it may be invovled in protein breakdown, to free more nitrogen for cellular use, due to its association with Srp1p. Srp1p is thought to be associated with the degredation of proteins. |
Future Experiments for YKL069W
After searching many proteomics databases, I have concluded that there is very little known about the YKL069W protein. I believe there are a number of tests that should be done in order to gain a better understanding of this non-annotated ORF and corresponding protein. I would first start by performing a Yeast Two Hybrid experiment to determine at a basic level the proteins that interact with YKL069W. From these proteins one could potentially make better hypotheses about YKL069W function. I would then suggest performing the ICAT method. The ICAT method was created to determine the difference in protein quantity at different times during the cell cycle. The ICAT method works by taking two populations of yeast cells at different times during the cell cycle and labeling the two populations with different ICAT reagents. One of the two will be treated with the Heavy reagent and one with the Light reagent. From the different labeling one can determine amount of protein present at different times during the cell cycle. This should help determine the function by narrowing the time when YKL069W is most active in the cells. The next step I would take is to design an YKL069W yeast knockout. This will provide information regarding the function of this protein. At the very least will allow one to determine if the protein is necessary for viability. By knocking out the YKL069W gene, one should be able to determine through a battery of tests (possibly growth on various limited mediums) which functions in the cell are not being performed at the same efficiency or not being performed at all. It is possible that an homologous protein could be present in the cell and a loss of function would not be seen. The final experiment that I would suggest. Would be an attempt to crystallize the YKL069W protein. Through X-ray crystallography one can determine a more accurate representation of the structure of the protein inside of the cell. Knowing the precise structure may allow individuals to more accurately determine the function of the protein. Even though there is a crystallized structure of a synthetic protein (which happens to have the same amino acid sequence) it would still be interesting to note the difference between the synthetic version of YKL069W and the actual protein isolated from living cells. Overall there are many experiments that should be done to better characterize YKL069W, however this is a slow process and will probably not be done anytime in the near future. Many of the experiments still need to be performed on proteins of known function. |
References:
DIP Database of interacting proteins. 2004. Last accessed November 17 2005.
Dolinski K., et. al. 2004. Saccharomyces Genome Database. Last accessed November 17 2005.
[KEGG] Kyoto Encyclopedia of Genes and Genomes http://www.genome.jp/kegg/genes.html Last accessed November 16 2005
Martinez SE, Beavo JA, Hol WGJ. 2002 September. GAF Domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Molecular Interventions 2(5): 317-323.
[MIPS] comprehensive yeast genome database. Last accessed November 17 2005.
[PDB] Protein Data Bank. Last accessed November 17 2005.
[PROWL] Protein Info. Last accessed November 17 2005.
Schwikowski B, et al. 2000. A Network of Protein-Protein Interactions in Yeast. Nature Biotechnology 18: 1257-1261.
[TRIPLES] TRansposon-Insertion Phenotypes, Localization, and Expression in Saccharomyces. Last accessed November 17 2005.
Uetz P, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623-7.
Zoraghi R, Corbin JD, Francis SH. 2004. Properties and Functions of GAF Domains in Cyclic Nucleotide Phosphodiesterases and Other Proteins. Molecular Pharmacology 65:267-278.
Any questions or concerns regarding this page should be directed to magemberling "at" davidson.edu.