This webpage was produced as an assignment for an undergraduate course at Davidson College.
Assignment 1: Publication Review
Whole-genome sequencing of 16 different mosquito species reveal rapid evolution and could inform malaria research

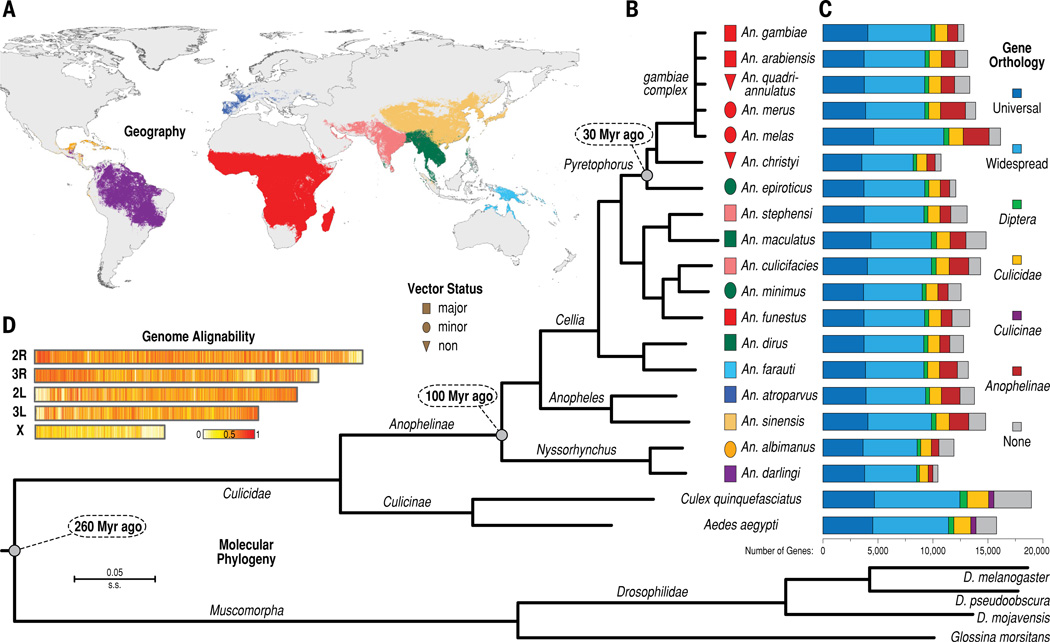
The phylogenetic tree (1B) generated by Neafsey et al. shows the evolutionary basis (or rather, lack thereof) for malaria vectorial capacity. Squares indicated major vectors, ellipses indicate minor vectors, and triangles indicate non-vectors. As demonstrated above, major and minor vectors did not deviate from non-vectors at any given point; rather, some major vectors are more closely related to non-vectors than they are to other major vectors. This figure suggests that vector status is determined by genes that were exchanged during interspecies mating, rather than inherited from a common ancestry. Published with permission from Neafsey et al.
Project Summary
In
two
papers published in Science on
November 27, 2014, researchers sequenced the genomes of sixteen species of
mosquitoes, some of which are known malaria vectors. In investigating the
genomic differences between mosquito species, researchers were able to
identify what genes dictate vectorial capacity for malaria, a disease that
affects millions of people annually. One paper detailed the process of
sequencing assorted mosquito species and their closest relatives to create
a phylogenetic tree representing the evolution of the mosquito vectors.
Interestingly enough, they found that vectorial capacity did not evolve at
once; rather, vector species are on distant branches of the evolutionary
tree.
Hypothesis or
discovery science?
The
genomic
research performed by the team at University of Notre Dame constitutes
discovery science. The team did not propose a hypothesis beyond suggesting
that there may be a genomic basis on top of previously known factors that
determine mosquitoes’ vectorial capacity. Otherwise, the sequencing,
comparative analyses, and creation of phylogenetic trees were purely
investigative.
Methods and
Genomic Technologies
Researchers
assembled
the genomes and transcriptomes of sixteen mosquito species (both lab and
wild specimens) by using Illumina sequencing technologies on genomic DNA
and whole-body RNA from each specimen. Sequences were annotated with MAKER
and the resulting gene count was satisfactory, with some variation of
total counts due to differing levels of assembly contiguity (Neafsey
et
al. 2014). Maximum-likelihood phylogenies were constructed
with various root species (which did not alter findings) and any
discordance between trees was resolved by implementing more in-depth
sequence analysis as detailed in the original publication (Fontaine
et al. 2014).
Key Points
Being
able
to identify the genes responsible for malaria vector status is a big first
step in implementing new technologies for malaria control. The
computational genome comparisons between the sixteen species identified
rapid evolution in mosquitoes, including high rates of gene gain, loss,
rearrangement, and interspecies transmission. The evolutionary trees that
the research team generated showed that vector species are present on
distant branches, and thus, did not evolve separately from non-vector
species. This leads researchers to hypothesize that the main factor in
vector status is introgression. Understanding this connection between
malaria vectors will allow for a strategized disease control
that more effectively targets malaria vectors specifically.
Evaluation
While the two publications summarized in this article
did not make conclusive statements about a specific gene or set of genes
that connect the malaria vector species, they have taken big steps in
sequencing such a large number of species and in finding that vector
status was not developed by the divergence of two ancestral species but
rather by introgression. Their findings, which indicate unusually high
rates of gene gain and loss, confirm the researchers' speculations about a
genomic factor at play in determining vectorial capacity. Their research
was purely investigative, and their findings suggest many more possible
directions for research. Since malaria is a disease that affects
populations not only in Africa, but in South America and Asia as well,
it’s essential that research move beyond studying just the principal
vector in Africa. Additionally, taking a multiple species sequencing
approach can establish a precedent for future research into other
vector-borne (and specifically, mosquito-borne) diseases, such as Dengue
fever, Yellow fever, and West Nile virus. It will be interesting to see
whether with the interspecies and vector/non-vector comparisons that this
research provides, future investigators can pinpoint a particular genomic
difference that determines vectorial capacity, and whether that genomic
difference can extend to other non-insect vectors. Approaching malaria and
other disease control from a genomic approach has potential to be more
efficient, cost-effective, healthy, and environmentally friendly than
blanket fumigation of at-risk malaria areas.
Citations
Fontaine,
Michael
C. et al. “Extensive
Introgression in a Malaria Vector Species Complex Revealed by
Phylogenomics.” Science (New York, N.Y.) 347.6217 (2015): 1258524.
PMC. Web. 25 Jan. 2016.
Neafsey,
Daniel
E. et al. “Highly
Evolvable Malaria Vectors: The Genomes of 16 Anopheles Mosquitoes."
Science (New York, N.Y.) 347.6217 (2015): 1258522. PMC. Web. 25 Jan.
2016.
Williams,
Ruth. "Mosquito Genomes Galore." The Scientist. 27 Nov. 2014. Retrieved
from http://www.the-scientist.com/?articles.view/articleNo/41546/title/Mosquito-Genomes-Galore/.
Please send comments and questions to Dylan Maghini at dymaghini@davidson.edu
© Copyright 2016 Department of Biology, Davidson College, Davidson, NC 28035