
photo from: http://tomatomania.com/
This web page was produced as an assignment for an undergraduate course at Davidson College.
Article: "A chemical genetic roadmap to improved tomato flavor"
Commercially grown tomatoes are available throughout the year and across the world. For this to be possible, tomato farmers have continually selectively breeded for tomatoes demonstrating firmness, high yield, and disease resistance. An implied result of this breeding is the loss of flavorful components over time, leaving us with durable, but rather mundane tasting tomatoes. Researchers utilized tasting panels, chemical analyses, and a genome wide association study of 398 varieties of tomatoes in order to identify genes associated with levels and types of sugars, acids, and volatiles contributing to tomato flavor (volatiles associated with olfactory reception -smell- which we know contributes to taste).
The study was developed upon the realization that commercially grown tomatoes lacked flavor compared to heirloom varieties. However, researchers had no hypothesis on the relationship between certain chemical compounds and tomato flavor. Thus, this was discovery science as researchers compared the sequenced genomes of tomato varieties with their taste and chemical testing in order to identify any genes that could be associated with rich flavor.
This study used a genome wide
association study (GWAS). This means that they sequenced the genomes of
398 varieties of tomatoes and then compared the genome sequences in order
to identify key similarities and differences that were related to the
existence of certain flavor-associated phenotypes. A key function of a
GWAS is to identify single nucleotide polymorphisms (SNPs), or variations
in a single base pair of the genome. In this study, researchers aimed to
uncover SNPs with a high likelihood of impacting sugar, acid and volatile
compound presence. This study targeted SNPs in the exome, or
"protein-coding region" of the genome. Thus, SNPs could either alter the
amino acid sequence of a protein by exchanging one amino acid with
another, truncate a protein by adding a premature stop codon, or shift the
reading frame of a protein by inserting or deleting bases in a multiple
other than 3 (called an indel). In each case, the amino acid sequence of a
protein would be altered, therefore changing a proteins functionality or
disabling it entirely.
For more info on genome wide association studies, check this out: https://www.genome.gov/20019523/
The "take home message of the study was that by comparing the exomes of 398 varieties of tomato, researchers were able to identify genes that were likely contributing to the reduction in flavor in commercial tomatoes. In order to make this determination, researchers not only identified differences in the genome, but thoroughly researched the wild-type and mutant proteins involved in order to assess the impact of these variations and prioritize a few genes with a strong likelihood of involvement in tomato flavor reduction.
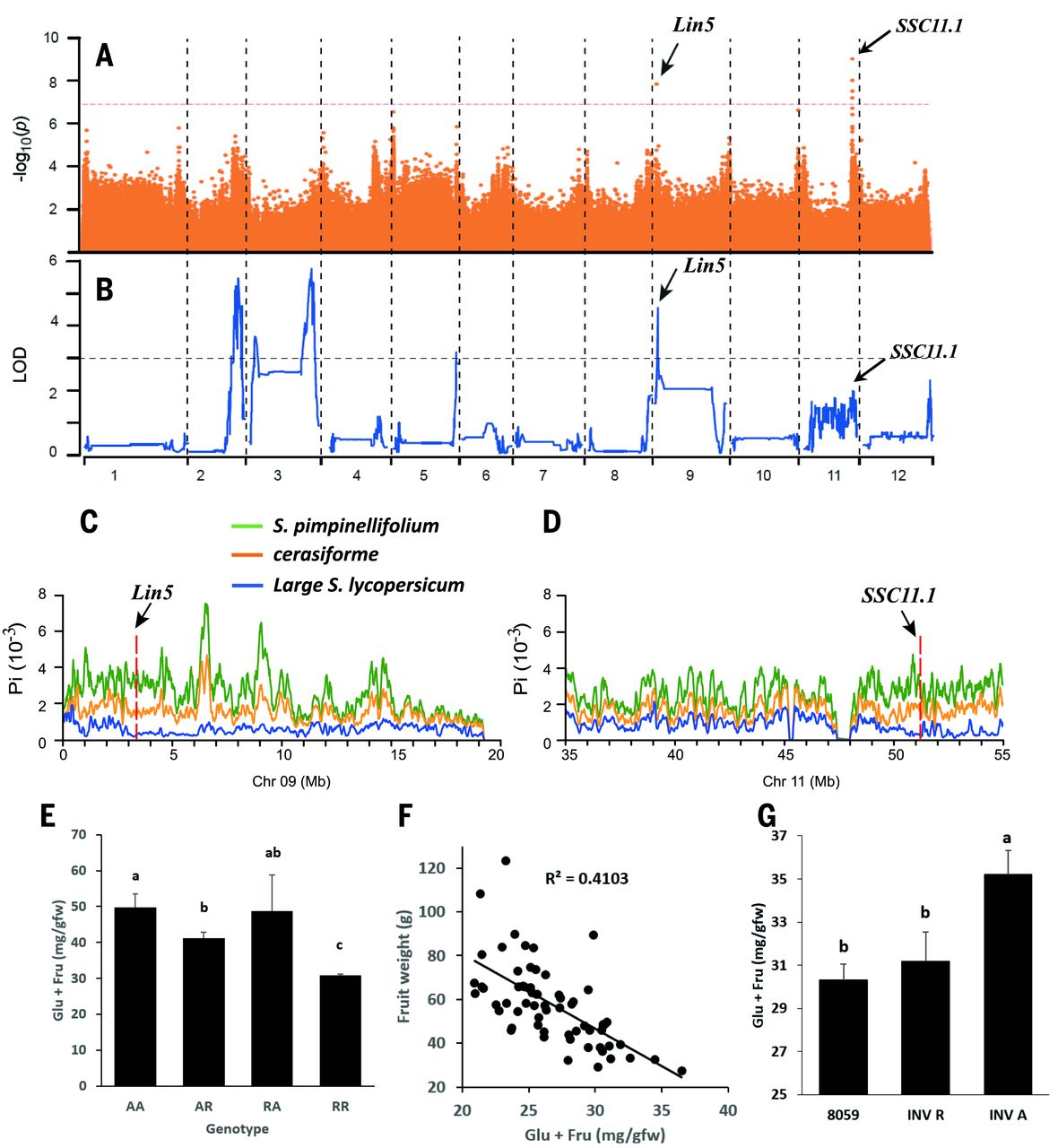
I felt that the project was thorough. The consideration of flavor-associated compounds being not only sugars and acids but also "volitiles" that give certain scents that contribute to taste I felt was very insightful. In addition, the combination of GWAS and its required data mining with targeted, hypothesis-driven experimentation was very effective in demonstrating not only the result in relation to tomatoes but also the power of GWAS and the use of "discovery science" assuming n (sample size) is large enough. One thing I would change is in figure 1G, sample sizes used to generate the data for reference (R) and alternate (A) were 6 and 17, respectively. I am curious as to why researchers did not use equal sample sizes, as tomatoes are easy enough to acquire. One possibility is that using these sample sizes gave researchers the most persuasive data, and that equal, larger sample sizes may not have been as effective in demonstrating their point. Due to the discrepancy, it may be worthwhile to be skeptical of the results of this figure.
References:
Denise Tieman et al. Jan 2017 : 391-394. "A Chemical Genetic Roadmap to Improved Tomato Flavor." A Chemical Genetic Roadmap to Improved Tomato Flavor | Science. Science, 27 Jan. 2017. Web. 29 Jan. 2017.
Return to Nick Balanda's Home Page
Email Questions or Comments: nibalanda@davidson.edu